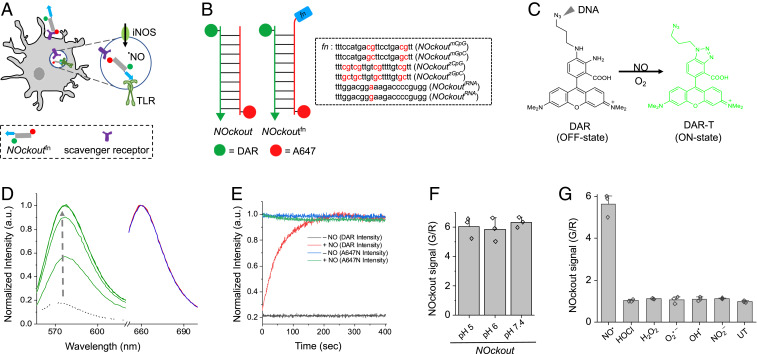
Fig. 1.
Design, sensitivity, and specificity of NOckout probes. (A) Schematic of TLR-mediated NOS2 activation by NOckoutFn. NOckoutFn, displaying a functional PAMP (Fn, blue), is phagocytosed and binds its cognate TLR-receptor (green hook) to activate NOS2 (green circle) and elevate phagosomal NO levels. (B) Schematic of the design of NOckout and NOckoutFn probes. NOckout is assembled from an ssDNA (24-mer) carrying an NO-sensing fluorophore (DAR, green circle) and a cDNA strand displaying a normalizing fluorophore (A647, red circle). NOckoutFn carries a functional (Fn) immunostimulatory CpG DNA or RNA motif as an overhang specific to mouse (NOckoutmCpG or NOckoutiRNA) or zebrafish TLR receptors (NOckoutzCpG or NOckoutiRNA). (C) Reaction with NO turns the nonfluorescent DAR probe (off-state) on NOckout into the highly fluorescent DAR-T (on-state). (D) Fluorescence spectra of NOckout (250 nM) in sodium phosphate buffer (pH 6.0, 50 mM) recorded in the DAR channel (green, λex = 550 nm, λem = 575 nm) and in the A647 channel (λex = 645 nm, λem = 660 nm) before (red) and after (blue) the addition of diethylamine NONOate (DEA NONOate, 50 μM, pH 6.0). Spectra in the DAR channel (G) were recorded in 30-s intervals (green traces). (E) Representative kinetic trace of NOckout in the DAR channel with (red) and without (black) DEA NONOate (50 μM, pH 6.0). Intensity in A647 channel before and after addition of DEA NONOate is shown in blue and green, respectively. (F) Fold change of NOckout represented as G/R values in pH 5, pH 6, and pH 7 buffers. (G) In vitro specificity of NOckout against various ROS and RNS. NOckout (250 nM, pH 6.0) was incubated with NO (50 μM of DEA NONOate), HOCl (5 μM), H2O2 (100 μM), O2.− (100 μM), OH· (100 μM), and NO2− (100 μM) for 6 min at room temperature, and the intensities were plotted as (G/R) values corresponding to DAR (G) to that of A647 (R). Error bars are SDs for n = 3 independent trials.