Significance
Extracting information regarding calendar time from seasonal changes in photoperiod and temperature is critical for organisms to maintain annual cycles in physiology and behavior. Here we found that, in flies, EYES ABSENT (EYA) protein acts as a seasonal sensor by adjusting its abundance and phase in response to changes in photoperiod and temperature. We show that the manipulation of EYA levels is sufficient to impair the ability of female Drosophila to regulate seasonal variation in reproductive dormancy. Finally, our results suggest an important role for the circadian clock protein TIMELESS (TIM) in modulating EYA level through its ability to measure night length, linking the circadian clock to seasonal timing.
Keywords: photoperiod, temperature, seasonality, alternative splicing, circadian clock
Abstract
Organisms possess photoperiodic timing mechanisms to detect variations in day length and temperature as the seasons progress. The nature of the molecular mechanisms interpreting and signaling these environmental changes to elicit downstream neuroendocrine and physiological responses are just starting to emerge. Here, we demonstrate that, in Drosophila melanogaster, EYES ABSENT (EYA) acts as a seasonal sensor by interpreting photoperiodic and temperature changes to trigger appropriate physiological responses. We observed that tissue-specific genetic manipulation of eya expression is sufficient to disrupt the ability of flies to sense seasonal cues, thereby altering the extent of female reproductive dormancy. Specifically, we observed that EYA proteins, which peak at night in short photoperiod and accumulate at higher levels in the cold, promote reproductive dormancy in female D. melanogaster. Furthermore, we provide evidence indicating that the role of EYA in photoperiodism and temperature sensing is aided by the stabilizing action of the light-sensitive circadian clock protein TIMELESS (TIM). We postulate that increased stability and level of TIM at night under short photoperiod together with the production of cold-induced and light-insensitive TIM isoforms facilitate EYA accumulation in winter conditions. This is supported by our observations that tim null mutants exhibit reduced incidence of reproductive dormancy in simulated winter conditions, while flies overexpressing tim show an increased incidence of reproductive dormancy even in long photoperiod.
As in plants and other animals, insects possess endogenous photoperiodic timers, which prompt them to undergo physiological and behavioral changes to survive through unfavorable periods. Arguably, the most well-recognized seasonal response in insects is the induction of overwintering diapause. This phenomenon can be induced at different life stages and is characterized by a reversible arrest in growth and/or reproduction in response to decreasing day length. Since photoperiodic time measurement (PPTM) is critical to seasonal adaptation in insects, it has been studied extensively (1, 2). Yet, the molecular and neuronal basis of the insect photoperiodic timer remains poorly understood.
Extensively studied for its role in eye development (3, 4), the EYES ABSENT (EYA) protein represents a promising target to unravel the molecular mechanisms underlying PPTM. This cotranscription factor with phosphatase activity is highly conserved in the animal kingdom (5) and has been implicated in diverse biological processes such as organ development, innate immunity, DNA damage repair, angiogenesis, and cancer metastasis (6–10). Interestingly, the mammalian ortholog, EYES ABSENT 3 (EYA3), was recently implicated in sheep as a clock-regulated transcription factor in the pituitary gland to promote the transcription of TSHβ, leading to an increase in thyroid-stimulating hormone and an induction of summer phenotypes in long photoperiod (11–15). This elegant work suggests that EYA3 plays a central role in the photoperiodic switch synchronizing circannual rhythms in reproduction with the environment. Nonetheless, due to inherent limitation of using sheep as animal subjects for those studies and the pleiotropic functions of EYA during development, there is still a lack of in vivo functional data from genetic manipulation of eya expression to confirm this hypothesis. Here we take advantage of the versatile genetic tools available in Drosophila melanogaster to investigate the role of EYA in insect photoperiodism.
Unlike many insect species relying mainly on photoperiodic signal to overwinter (16), D. melanogaster requires cold temperature to enhance reproductive dormancy under short photoperiod (17). Whether D. melanogaster represents a suitable model to study diapause has been under debate. However, recent studies suggest that the photoperiodic component of D. melanogaster might be more robust than previously described (18, 19). We found that newly emerged females reared at 10 °C for 28 d exhibit significantly smaller ovaries when exposed to short photoperiod (SP 8L:16D, where L denotes hours of light and D denotes hours of dark) compared to long photoperiod (LP 16L:8D). Using these conditions as readout for reproductive dormancy in combination with the inducible gene-switch driver to genetically manipulate eya expression in a spatiotemporal manner (3, 20–22), we provide functional evidence for the role of EYA in insect PPTM, specifically, in promoting winter physiology. We are referring to the phenomenon of ovary growth arrest in D. melanogaster as reproductive dormancy rather than diapause, due to its reliance on temperature in addition to photoperiodic cues.
Finally, we present results suggesting that the function of EYA in seasonal adaptation is aided by cold temperature-dependent induction of light-insensitive TIMELESS (TIM) isoforms, which contribute to EYA stabilization in winter condition. Our results directly link a key circadian clock protein to insect seasonal timing.
Results
Reproductive Dormancy Is a Robust Phenotypic Readout for Winter Physiology in D. melanogaster.
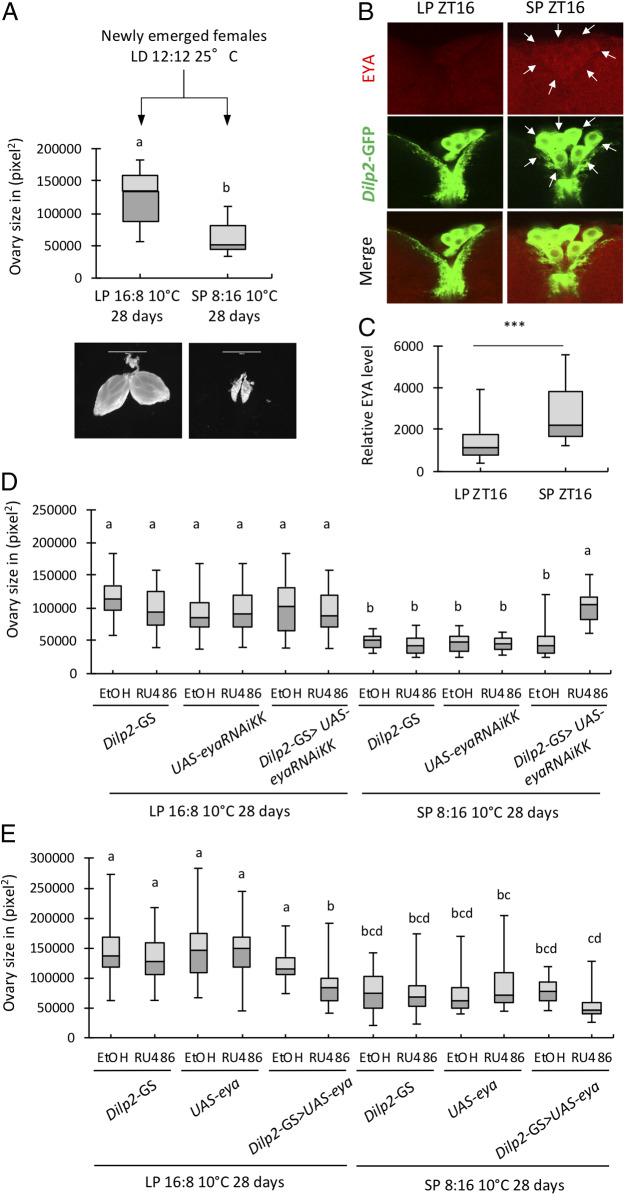
To verify that reproductive dormancy (i.e., ovary size) in D. melanogaster can serve as a robust phenotypic readout for long photoperiod (LP 16L:8D) versus short photoperiod (SP 8L:16D), we subjected wild-type (WT) (w1118) flies to a reproductive dormancy assay in which they were reared for 28 d at 10 °C in either LP or SP. Under SP, flies showed significantly smaller ovary size when compared to flies maintained in LP (Fig. 1A). This provides a suitable readout to investigate the effect of eya genetic manipulation in the photoperiodic control of seasonal physiology. As shown in previous studies (18), we found that temperature above 10 °C to 12 °C was not sufficient to induce D. melanogaster reproductive dormancy in SP.
Fig. 1.
Genetic manipulation of eya in the PI impacts reproductive dormancy. (A) Levels of reproductive dormancy determined by ovary size measurement (in pixel2) in WT (w1118) females reared for 28 d at 10 °C in long photoperiod (LP 16L:8D) and short photoperiod (SP 8L: 16D). The whisker caps represent the minimum and maximum values; different letters indicate significant differences in ovary size between groups. All error bars indicate SEM. Mann−Whitney test, P < 0.0001, n = 40. (Scale bar: 1,000 μm.) (B) Comparison of EYA levels in dilp2-Gal4 > UAS-cd8-GFP brains between LP and SP at ZT16 (10 °C). White arrows denote EYA-positive cells. (Magnification: 355×.) (C) Quantification of EYA staining in IPCs. Five or six neurons per brain were imaged for eight or nine brains. ***P < 0.001, Mann−Whitney test. Ovary size of females (D) expressing eya dsRNAs (dilp2-GS > eyaRNAiKK in the presence of RU486) or (E) overexpressing eya (dilp2-GS > UAS-eya in the presence of RU486) in dilp2 neurons at adult stage as compared to parental controls and vehicle control (EtOH) in LP and SP. Kruskall−Wallis test with Dunn’s multiple comparison test, P < 0.001, n= 30 to 40 per group.
To ensure that the observed differences in reproductive dormancy are primarily driven by photoperiod length and not heavily dependent on minor thermal variations associated with duration of light period in LP vs. SP, we reduced temperature variations between light and dark periods to ∼ ±0.25 °C by adjusting incubator temperature settings, and evaluated reproductive dormancy in WT flies (SI Appendix, Fig. S1A). A previous study found that even temperature cycling of ±0.3 °C for 12 d can generate an apparent LP vs. SP effect on reproductive dormancy (18). In LP, WT females exhibited significantly smaller ovaries when temperature variations between light and dark periods were minimized (SI Appendix, Fig. S1B). Most importantly, WT ovaries in SP with reduced temperature variations between light and dark periods were still significantly smaller than the corresponding ovaries in LP. Since we were unable to completely abolish the temperature variations between light and dark periods, we cannot rule out the possibility that thermal differences contributed to the larger ovaries we observed in LP. Nevertheless, it should be noted that there is no light without heat in nature.
eya Mediates Photoperiodic Regulation of Reproductive Dormancy.
To lay the groundwork for tissue-specific manipulation of eya expression, we performed immunostaining in adult fly brains by driving green fluorescent protein (GFP) expression under eya promoter. We observed GFP expression in the optic lobes at ZT8 (LD12:12 at 25 °C) (SI Appendix, Fig. S2). We also observed GFP expression in insulin-producing cells (IPCs) within the pars intercerebralis (PI) and dorsal lateral peptidergic (DLPs) neurons located in the pars lateralis (PL). Expression pattern of eya correlates with its confirmed role in eye development as well as its potential role in regulating neuroendocrinology of seasonality. To confirm EYA expression in IPCs, we performed EYA and GFP double staining in dilp2-Gal4 > UAS-GFP fly brains and compared EYA levels between LP and SP (ZT16) at 10 °C. The choice of ZT16 was justified by EYA protein analysis described later in this study. Our results clearly indicated the presence of EYA in IPCs. In addition, we observed significantly higher EYA signal in dilp2-expressing neurons under SP (Fig. 1 B and C), consistent with the potential role of EYA in promoting reproductive dormancy under SP.
We first tested the requirement of eya expression in the PI for reproductive dormancy, as it has been identified as an important brain region for neuroendocrine control of insect physiology. In particular, insulin signaling has been shown to be an important regulator of seasonal adaptation (23–27). Dilp2-GS gene-switch Gal4 driver (28) was used to drive the expression of eya double-stranded RNAs (dsRNAs) in the IPCs within the PI at the adult stage to knock down eya in a time- and tissue-specific manner to limit pleiotropic and developmental effects (Fig. 1D). Reduction in eya expression has no effect on ovary size in LP at 10 °C but led to a significant increase in ovary size in SP at 10 °C when compared to all control groups. Conversely, when we overexpressed eya in dilp2-expressing neurons at the adult stage, females showed significant decrease in ovary size when compared to parental controls and vehicle control in LP at 10 °C (Fig. 1E). The ovaries of eya overexpressors in LP were comparable to that observed in flies reared in SP.
We also tested the importance of eya expression in gmr-expressing cells in the visual system (SI Appendix, Fig. S3 A and B), since we observed prominent eya expression in the optic lobes (SI Appendix, Fig. S2). Knockdown of eya using gmr-GS gene-switch driver (29) at the adult stage similarly resulted in significant increase in ovary size specifically in SP at 10 °C (SI Appendix, Fig. S3A). On the other hand, eya overexpression in gmr-expressing cells led to significant decrease in ovary size even in LP (SI Appendix, Fig. S3B). These observations are consistent with our results using eya-Gal4 driver to broadly overexpress eya (SI Appendix, Figs. S1C and S3C). Our results provide functional evidence supporting the involvement of eya in the regulation of reproductive dormancy in arthropods.
EYA Senses Both Photoperiod and Temperature.
Since photoperiodic signal constitutes a major predictable cue to anticipate seasonal progression, we hypothesize that eya may interpret photoperiod length and modulate its expression at transcriptional and/or protein level to regulate seasonal physiology. To test this hypothesis, we evaluated daily eya messenger RNA (mRNA) and EYA protein expression from head extracts of WT (w1118) flies before and after application of a photoperiodic shift, by rearing flies for 3 d in LP followed by 7 d in SP (Fig. 2A). Temperature was set at 10 °C throughout the entire experiment after fly emergence, in order to maintain conditions necessary for reproductive dormancy and specifically assess the contribution of photoperiod. Samples were collected at days 3, 5, and 10 (LP3, SP5, and SP10, respectively) and assayed by qPCR and Western blotting (Fig. 2 B–G). The eya mRNA and proteins were analyzed at multiple time points over a 24-h light−dark cycle, as mammalian Eya3 mRNA was shown to be clock regulated (30). Analysis of EYA proteins on LP3, SP5, and SP10 showed robust oscillation over the daily cycle in all three groups. A striking effect of the photoperiodic shift was an 8-h delay in peak phase at the protein level between LP (ZT8) and SP (ZT16). This corresponds precisely to the difference in day length between the respective photoperiodic regimes. Unlike EYA proteins, the phase of eya mRNA oscillations remains unchanged between LP and SP conditions (Fig. 2 E–G). This observation suggests a possible role of EYA in sensing day length at the protein level, specifically, peaking in the dark phase in SP. The delay in protein relative to the mRNA peak suggests that posttranscriptional mechanisms may be important in shaping EYA protein profile.
Fig. 2.
The peak phase of EYA protein expression is regulated by photoperiod. (A) Schematic diagram illustrating conditions and collection time points for photoperiodic shift experiments (black arrows denote collection time points). Samples were collected on LP3, SP5, and SP10 at 4-h intervals over 24-h periods. (B–D) Western blots and quantifications comparing EYA expression profiles between head extracts of w1118 on LP3, SP5, and SP10 (peak phase: LP3 = ZT8, SP5 and SP10 = ZT16, P < 0.01 for all conditions, JTK-CYCLE). EYA protein levels were detected using ⍺-EYA10H6 (top isoform was used for quantification). ⍺-HSP70 was used to indicate equal loading and for normalization. (E–G) Comparison of eya mRNA expression in heads of w1118 flies between LP3, SP5, and SP10 normalized to cbp20 (peak phase: all at ZT12, P < 0.05 for all conditions, JTK-CYCLE). The gray shading on each graph indicates when lights were off during each sampling period (ZT: Zeitgeber time [hours]). Data are mean ± SEM of n = 4 replicates for mRNA analysis (two technical replicates for each of the two biological replicates) and n = 3 biological replicates for protein analysis.
Interestingly, we observed a significant increase between SP5 and SP10 in peak eya mRNA expression at ZT12 (1.51-fold change) and EYA protein at ZT16 (1.67-fold change), respectively (Fig. 2; P < 0.05, two-way ANOVA with post hoc Tukey’s honestly significant difference [HSD] tests). This led to the hypothesis that a prolonged exposure of SP, perhaps in combination with low temperature, might be responsible for the higher level of eya mRNA and EYA protein on SP10. To test this hypothesis, flies were reared in light−dark cycle (LD 12L:12D) for 3 d at 25 °C (3D25) before being transferred to either 10 °C (10D10) or kept at 25 °C (10D25) for another 7 d (Fig. 3A). As expected, we observed an increase between 10D25 and 10D10 with a 1.70- and 1.44-fold change in eya transcript at ZT12 (P < 0.05, two-way ANOVA with post hoc Tukey’s HSD tests) and EYA protein at ZT16 (P < 0.001), respectively (Fig. 3 B–G). This supports a model in which eya is a cold-induced gene in D. melanogaster and the cold induction of EYA protein can at least be partially attributed to induction at the transcriptional level.
Fig. 3.
Low temperature induces significant increase in eya expression and amplitude at both transcriptional and protein levels. (A) Schematic diagram depicting experimental conditions for testing effect of temperature on eya mRNA and EYA protein. Newly emerged flies were reared at 10 °C for 3 d in LD12:12 at 25 °C followed by 7 d at either 25 °C or 10 °C. Flies were collected on day 3 prior to shifting half of the flies into 10 °C and on day 10 at 4-h intervals over 24-h periods. (B−D) Western blots and quantifications comparing EYA expression profiles between head extracts of w1118 on 3 d at 25 °C, 10 d at 25 °C, and 10 d at 10 °C (peak phase: 3D25 = ZT14, 10D25 = ZT16, 10D10 = ZT14, P < 0.0001 for all conditions, JTK-CYCLE). EYA levels were detected using ⍺-EYA10H6 (top isoform was used for quantification). ⍺-HSP70 was used to indicate equal loading and for normalization. (E–G) Comparison of eya mRNA expression in heads of w1118 flies between 3 d at 25 °C, 10 d at 25 °C and 10 d at 10 °C normalized to cbp20 (peak phase: all at ZT12, P < 0.05 for all conditions, JTK-CYCLE). The gray shading in each graph indicates when lights were off during each sampling period. Data are mean ± SEM of n = 4 for mRNA analysis (two technical replicates for each of the two biological replicates) and n = 3 biological replicates for protein analysis.
EYA Rhythmic Expression Is Not Directly Controlled by the Circadian Clock.
To determine whether eya daily oscillation is regulated by the circadian clock, we examined its expression in WT (w1118) flies in free-running condition and in Clk mutants (clkout) using qPCR. Although eya mRNA is rhythmic in LD cycles, these oscillations do not persist in constant darkness (DD) (Fig. 4A). This suggests that eya rhythmic expression may be regulated through a light-dependent pathway rather than being clock controlled. This result is supported by the fact that no significant change in eya mRNA expression and oscillation was observed in clkout mutants (Fig. 4B).
Fig. 4.
Light affects EYA protein stability. (A) The eya mRNA expression in heads of w1118 flies collected on LD4 and DD1 after 3 d of entrainment (LD1-3) at LD12:12 at 25 °C. (LD: peak phase = ZT12, p(LD) < 0.0001; p(DD) = 1, JTK-CYCLE). (B) The eya mRNA expression in heads of w1118 compared to w; clkout in LD (12:12). Flies were harvested on LD4 at LD12:12 at 25 °C (peak = ZT12, p(WT) < 0.0001, p(clkout) < 0.005, JTK-CYCLE). (C and D) The eya mRNA expression in heads of w1118 flies collected on (C) LD4 and LL1 (peak = ZT12, p(LD4) < 0.0001, p(LL1) < 0.001, JTK-CYCLE) and (D) LD5 and LL2 (peak = ZT12, p(LD5) < 0.0001; p(LL2) = 1, JTK-CYCLE) after 3 d of entrainment at LD12:12 at 25 °C. Data are mean ± SEM of n = 4 (two technical replicates for each of the two biological replicates). Asterisks denote significant differences between conditions or genotypes at each ZT: ***P < 0.001, *P < 0.05, two-way ANOVA with post hoc Tukey’s HSD tests. (E) Western blots comparing EYA expression profiles in heads of w1118 flies collected on LD4 and LL1. Top, Middle, and Bottom panels detect EYA, TIM, and HSP70 expression, respectively. (F) Quantification of E and expressed as relative expression (highest value = 1) (peak = ZT16, p(LD4) < 0.005; p(LL1) = 1, JTK-CYCLE). Second replicate of protein analysis is shown in SI Appendix, Fig. S4A (n = 2).
To further examine the role of light in the regulation of eya mRNA, we compared expression of eya in WT flies in LD to those in constant light condition (LL1 to LL2) (Fig. 4 C and D). The eya daily mRNA expression remains unaffected in LL1 compared to LD4 after 3 d of LD entrainment (Fig. 4C). However, when comparing LD5 and LL2, eya mRNA abundance was significantly reduced in LL2 at ZT8, ZT12, and ZT16, and no oscillation was detected. Interestingly, EYA protein level was dramatically reduced even on LL1 as compared to LD4, and the oscillation was dampened (Fig. 4 E and F and SI Appendix, Fig. S4A). The effect of light on EYA protein stability shows striking similarities with the well-characterized light-mediated TIM degradation (31–33) (Fig. 4 E, Middle). One difference between EYA and TIM expression in LD is that EYA appears to accumulate at earlier time points compared to TIM, which shows an expression pattern restricted to the dark phase only.
Genetic Manipulation of tim Alters EYA Stability and Reproductive Dormancy.
Since EYA protein exhibits a pattern similar to TIM protein under LL, we decided to examine its expression level in tim mutant flies to investigate whether TIM is involved in stabilizing EYA. Genetic variations in tim have previously been linked to latitudinal clines in diapause incidence in Drosophila (34, 35). By Western blotting, we observed that EYA abundance is markedly reduced in tim null mutants (yw; tim0) (Fig. 5 A and B and SI Appendix, Fig. S4B) and starts to accumulate after lights-off. Conversely, an increase in EYA was detected in tim-overexpressing flies (w; p{tim(WT)}) (Fig. 5 C and D and SI Appendix, Fig. S4C). Interestingly, no significant change in eya mRNA expression was detected when comparing WT to tim null mutants or to tim overexpressors at all time points (Fig. 5 E and F), supporting the role of TIM in stabilizing EYA at the protein level.
Fig. 5.
Genetic manipulation of tim affects EYA protein stability and incidence of reproductive dormancy. (A and C) Western blots comparing EYA expression profiles in tim null mutant (yw; tim0) and tim overexpressor (w1118; p{tim(WT)}) to WT (yw or w1118) controls. Flies were entrained in LD (12:12) at 25 °C for 3 d and collected at six time points on LD4. (B and D) Quantification of Western blots shown in A and C and expressed as relative expression (highest value = 1). Second replicates of protein analyses are shown in SI Appendix, Fig. S4 B and C (n = 2). (E and F) Daily eya mRNA expression in tim null mutant (yw; tim0) and tim overexpressor (w1118; p{tim(WT)}) as compared to respective WT controls. Data are mean ± SEM of n = 4 (two technical replicates for each of the two biological replicates). Two-way ANOVA with post hoc Tukey’s HSD tests. (G and H) Ovary size of female tim null (yw; tim0) and tim overexpressor (w; p{tim(WT)}) as compared to WT (yw or w1118) in reproductive dormancy assay. The whisker caps represent the minimum and maximum values; different letters indicate significant differences in ovary size between groups with P < 0.001, except for difference between b and c in H. P < 0.05, n = 40 females per group, one-way ANOVA followed by post hoc Tukey test.
We then proceeded to test the effect of tim genetic manipulations on female reproductive dormancy. We found that, when tim null mutants were reared in SP at 10 °C, their ability to enter into reproductive dormancy was significantly impaired as compared to WT (Fig. 5G). In a reciprocal experiment, we evaluated ovary size of tim-overexpressing flies in LP and SP and observed that the proportion of individuals in reproductive dormancy was significantly higher as compared to WT, even in LP (Fig. 5H). The difference in ovary size between WT and tim-overexpressing flies was also significant in SP but much smaller (Fig. 5H). Our results are consistent with the role of TIM in supporting EYA in the regulation of reproductive dormancy in response to photoperiodic signals.
TIM Mediates both Photoperiodic and Temperature Signals to Impact EYA Expression.
Although analysis of tim mutants supports the hypothesis that TIM may promote EYA expression, the low level of canonical TIM expression at low temperature (36) and its absence during the light phase (31–33) suggest that other TIM isoforms could be involved. We therefore evaluated whether the expression profiles of different TIM isoforms are affected by temperature shift in a fashion similar to EYA. At 10 °C, canonical TIM was barely detectable using our TIM antibody when compared to TIM levels at 25 °C, and no other isoforms were detected. Concomitantly, recent studies revealed that tim undergoes a thermosensitive alternative splicing controlling the relative abundance of various tim isoforms (37, 38). Consistent with our observation, the authors found lower levels of the canonical TIM protein at 18 °C and revealed the induction of two cold-specific splice forms (tim-cold and tim-short&cold). The antibody we used to detect TIM was generated using a C-terminal antigen that is absent in the shorter TIM-SC isoform. In order to detect TIM-SC, we used a TIM antibody generated against a N-terminal antigen (32) and repeated the analysis. This time, we observed a drastic switch between the canonical TIM-L and the cold-induced TIM-SC isoform between 25 °C and 10 °C (Fig. 6A and SI Appendix, Fig. S4D). TIM-L abundance and its cycling amplitude were notably reduced at 10 °C as compared to that observed at 25 °C (Fig. 6B). Conversely, TIM-SC isoforms showed an opposite trend, with a substantial increase in expression at 10 °C, whereas almost no signal was detected at 25 °C (Fig. 6C). TIM-SC was expressed at high levels at 10 °C and exhibited a weak oscillation; however, this isoform failed to reach significance for Jonckheeree–Terpstra–Kendall (JTK) cycling statistics, likely due to its stability in the presence of light.
Fig. 6.
Expression of TIM-L and TIM-SC is temperature dependent. Flies were reared using the same condition as in Fig. 3. (A) Western blots comparing TIM expression profiles between head extracts of w1118 collected on 3 d at 25 °C, 10 d at 25 °C, and 10 d at 10 °C. Both TIM-L and TIM-SC isoforms were detected using ⍺-TIM (a gift from M. Young’s laboratory, The Rockefeller University, New York, NY) (32, 68), and ⍺-HSP70 was used to indicate equal loading and for normalization. (B and C) Quantification of Western blot signals shown in A and expressed as relative expression (highest value = 1). (B) Peak = ZT16, p(3D25) < 0.0001, p(10D25) < 0.0001; p(10D10) = 1, JTK-CYCLE; (C) p(3D25) = 1, p(10D25) = 1, p(10D10) = 0.064, JTK-CYCLE. Second replicate of protein analysis is shown in SI Appendix, Fig. S4D (n = 2). (D–G) Comparison of tim-M, tim-L+M, tim-cold, and tim-sc mRNA expression profiles in heads of w1118 flies between 3 d at 25 °C, 10 d at 25 °C, and 10 d at 10 °C normalized to cbp20. Data are mean ± SEM of n = 4 (two technical replicates for each of the two biological replicates). Asterisks denote significant differences between 10 d at 25 °C and 10 d at 10 °C at each ZT: ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05, two-way ANOVA with post hoc Tukey’s HSD tests.
The evaluation of the mRNA levels for different tim isoforms confirmed that both tim-cold and tim-sc are induced at 10 °C compared to 25 °C (Fig. 6 F and G), while tim-M and tim-L+M are drastically reduced in the cold (Fig. 6 D and E). All together, these results support the hypothesis that tim thermosensitive alternative splicing plays an important role in stabilizing EYA in cold temperature.
We also investigated whether expression of different tim isoforms are modulated by photoperiodic signals when temperature was kept at 10 °C. At the transcriptional level, we observed a significant reduction in tim-sc mRNA between ZT16 and ZT20 in response to photoperiodic shift from LP to SP at 10 °C (LP3 vs. SP10), whereas no significant changes were detected for the longer tim isoforms (tim-L+M, tim-M, and tim-cold) (SI Appendix, Fig. S5 A–D). In addition, we observed a 4-h phase advance in tim-sc expression peak as flies transitioned from LP to SP condition (SI Appendix, Fig. S5D). At the protein level, consistent with the effect of low temperature on canonical TIM protein, we observed relatively low levels of TIM-L across LP and SP conditions at 10 °C (SI Appendix, Figs. S4E and S5E). In SP5 and, even more so, in SP10, higher levels of TIM-L were observed at ZT0 and from ZT16 to ZT20 as compared to TIM-L at LP3 (SI Appendix, Fig. S5F). Robust daily cycling of TIM-L was detected in SP5 and SP10 at 10 °C, but not in LP3. Despite the effect of low temperature in diminishing TIM abundance, SP may still play a role in increasing TIM-L accumulation during nighttime, likely due to increased duration of the dark phase. With regard to TIM-SC, no change in peak phase was observed between LP and SP, although we detected a higher level of TIM-SC at ZT16 and ZT20 in SP10 as compared to LP3 (SI Appendix, Fig. S5G). Combining the results of temperature and photoperiodic shift experiments, SP in conjunction with low temperature provide the ideal condition for higher TIM-L and TIM-SC isoforms at night as well as elevated abundance of TIM-SC during the day. Both isoforms may function to stabilize EYA and promote reproductive dormancy. This is consistent with more abundant EYA and increased incidence of reproductive dormancy in flies overexpressing tim.
Finally, to test the potential of TIM in stabilizing EYA through protein interactions, we evaluated the ability of TIM-L and TIM-SC isoforms to bind EYA by expressing them in Drosophila S2 cells. Reciprocal coimmunoprecipitations suggested that both TIM isoforms can interact with EYA (Fig. 7A), although the affinity of TIM-SC to EYA is significantly higher as compared to TIM-L (Fig. 7B). To test whether EYA and TIM could colocalize in specific brain regions involved in photoperiodic sensing, we expressed GFP under the control of eya promoter and stained for TIM using the antibody that recognizes both TIM-L and TIM-SC isoforms. While we cannot rule out the possibility that we are observing some expression for TIM-L, it is reasonable to believe that most of the TIM signal observed corresponds to TIM-SC expression at 10 °C. We performed our analysis at ZT16 on SP5 since TIM and EYA expression was shown to be abundant by Western blot analysis. Analysis of whole-brain immunostaining revealed colocalization of eya and TIM in the optic lobes. While we detected signal for both eya and TIM in the PI, we did not find evidence for colocalization (Fig. 7 C–E).
Fig. 7.
TIM-SC shows higher affinity for EYA as compared to TIM-L. (A) Western blots showing results of reciprocal coimmunoprecipitation (coIP) assays to detect interaction of EYA with TIM-L and TIM-SC in Drosophila S2 cells. Proteins extracted from S2 cells were either immunoprecipitated with α-FLAG to pull down EYA or with α-HA to pull down either TIM-L or TIM-SC. Negative control coIPs were performed using α-V5, which do not recognize the proteins of interest. Immunocomplexes were subjected to Western blotting to detect the bait protein or protein interactions. Input for the coIP is indicated (Lys). (B) Bar graphs showing quantification of reciprocal coIP assays (n = 4). Values for binding of interacting proteins were normalized to amount of bait detected in the respective IPs. Error bars indicate ±SEM (*P < 0.05, two-tailed Student’s t test). (C–E) The eya-gal4/UAS-cd8-GFP brains from flies collected on SP5 (ZT16) at 10 °C were stained for (C) GFP and (D) TIM. OL: optic lobes; PI: pars intercerebralis. (E) Merged images showing colocalization of GFP (eya) and TIM in the OL but not in the PI. Eight or nine brains were imaged. Scale bars for whole brain and OL represent 100 nm and scale bar for PI represents 10 nm.
Discussion
In this study, we provide converging evidence supporting a central role of eya in seasonal adaptation in D. melanogaster (SI Appendix, Fig. S6). This includes immunocytochemical detection of EYA in areas within the fly brain regulating diapause and insulin signaling and the capacity of eya mRNA and protein to track temperature and photoperiod shifts through modulation of their expression. Perhaps the strongest evidence is our finding that spatiotemporal genetic manipulation of eya can alter seasonal physiology in the context of female reproductive dormancy. Our results support a conserved role of eya in regulating seasonal physiology in response to environmental cues in animals, but also highlight key differences between the mechanisms by which eya and Eya3 perform their functions in D. melanogaster and sheep.
In Drosophila, eya was first characterized as a key player for eye development and subsequently, more broadly, for organ development (8, 39). Our analysis of eya expression in the adult fly brain provides strong indication that eya functions beyond development. We observed that eya is expressed in specific regions within the brain: the PI and the PL, which house IPCs and DLPs neurons, respectively (40). Interestingly, these two clusters of neurosecretory cells play an important role in the hormonal control of insect diapause (25, 41). In a large number of insect species, it is well established that endocrine regulation of diapause in adult females is mediated by ILPs and Juvenile Hormone through the inactivation of the IPCs/copora allata/ovarian axis (23–27, 42, 43). In D. melanogaster, decreased IPC activity is associated with down-regulation of ilp2 and ilp3 production and correlates with an increased incidence of reproductive dormancy (26). It is possible that the function of EYA in photoperiodic sensing is at least partially mediated through insulin signaling.
In addition to neurosecretory cells, we also observed eya expression in adult optic lobes. Previous work demonstrated that the absence of compound eyes in clieya mutants affect their ability to entrain to extremely short or long photoperiods (44). The fact that knockdown of eya in the adult visual system can disrupt photoperiodic sensing indicates that EYA functions in transmitting photoperiodic signals in addition to its role in compound eye development. Compound eyes are necessary for phase adjustment, and, considering the important role of the visual system in light transduction (45), it is possible that EYA might play a role in mediating photoperiodic signals to other sites within the brain.
Although the use of D. melanogaster as a model to study insect photoperiodic timing has been historically challenging, there is accumulating evidence showing that adult reproductive dormancy in D. melanogaster can be induced by short photoperiod at sufficiently low temperature. A number of studies found that flies entrained in short photoperiod (<14 h of light per day) and low temperature (<12 °C) show significantly reduced vitellogenesis compared to females exposed to long photoperiod (16 h of light per day) at the same low temperature (43, 46, 47). In synergy with light, temperature plays a major role in Drosophila reproductive dormancy. Indeed, regardless of the photoperiodic conditions, flies entrained at 12 °C and above always undergo ovarian development. This is contrary to other insect species for which photoperiod plays a dominant role in the induction of seasonal phenotypes (48, 49). A key step of this study was therefore to establish a robust phenotypic readout for seasonal physiology. By rearing flies at 10 °C for 28 d after emergence either in LP and SP, we were able to induce significant phenotypic differences in reproductive dormancy, and used these conditions to evaluate the impact of manipulating eya expression in physiologically relevant conditions. We found that manipulation of eya in eya-expressing cells, and, more specifically, either in the visual system (gmr-expressing cells) or in insulin-producing neurons, had significant consequences on reproductive dormancy. Our results represent in vivo genetic manipulation data supporting a critical role of eya as a photoperiodic sensor in animals.
Another advantage of establishing robust conditions to induce reproductive dormancy in flies was the ability to track eya mRNA and protein expression in physiologically relevant conditions and investigate whether eya can “sense” photoperiodic and temperature shifts that ultimately result in different outcomes in reproductive dormancy. The mammalian ortholog Eya3 has been proposed to play an important role in photoperiodism (11, 14, 15). In sheep, Eya3 is a clock-controlled gene whose rhythmic expression is set by the phase of evening melatonin onset. This represents a classic example of external coincidence where light-dependent stimulus interacts with circadian oscillator to trigger seasonal phenotype (50, 51). We asked the question whether eya mRNA or protein can similarly interpret photoperiodic cues in insects. We observed that the phase of EYA oscillation is strongly influenced by photoperiod, while that for eya mRNA is not affected. Indeed, in LP, EYA peak expression is detected during the daytime, and transferring the flies into SP induces an 8-h shift in EYA peak phase, restricting its accumulation during the nighttime. In contrast, Dardente et al. (11) observed Eya3 photoperiodic shift occurring at the transcriptional level with an accumulation during daytime in LP, consistent with Eya3-dependent induction of a summer phenotype in sheep.
In mammals, the effect of changing day length on endocrine rhythm is mediated by nocturnal secretion of melatonin by suppressing the expression of a range of E box-controlled gene, including Eya3 (52). Melatonin has been implicated in photoperiodism in some insect species (53, 54); however, its involvement in Drosophila is largely debated. This could explain why we did not observe any significant variation in eya mRNA oscillation in response to changing photoperiods.
How eya is regulated at the transcriptional level in flies has not been resolved in this study. Despite the presence of E-boxes upstream of eya coding sequence (SI Appendix, Fig. S7), eya expression was unaltered in clkout mutant. In addition, daily cycling of eya mRNA was abolished in free-running condition, suggesting that eya transcription is not clock controlled. This is consistent with recent findings suggesting that photoperiodic sensing appears to be independent of robust circadian clocks (55). Among the potential pathways involved in regulating eya transcription, Pigment Dispersing Factor (PDF) signaling represents a strong candidate. PDF is well characterized for its role in photoperiodic sensitivity (56–62), and there is accumulating evidence supporting its involvement in reproductive dormancy by conveying day length signal to the PI (63, 64). We speculate that PDF could regulate eya expression in a cyclic adenosine monophosphate (cAMP)-dependent manner, perhaps in response to light signal. This is supported by the presence of CREB element in the eya promoter (SI Appendix, Fig. S7). Similarly in mammals, Eya3 transcription is regulated by cAMP (14).
Independent of the effect of light on eya mRNA expression, the abundance of EYA protein is significantly reduced under constant light, similar to the effect of light on TIM. To test our hypothesis that the light response of EYA is mediated by the light sensitivity of TIM and that TIM may stabilize EYA at the protein level, we evaluated EYA expression in tim mutants. As anticipated, we observed a correlation between EYA and TIM proteins with significantly higher EYA when TIM is abundant, and vice versa. In particular, under photoperiodic shift, we found that all TIM isoforms (TIM-L/TIM-Cold and TIM-SC) accumulate during the nighttime in SP10, matching the increase in EYA levels from ZT12 to ZT16 in the same conditions. Since this experiment was conducted at 10 °C, it is likely that the residual expression of the long TIM isoforms (TIM-L/TIM-Cold) we detected is, in most part, due to the induction of the TIM-Cold isoform. Nonetheless, due to their similarity in size, TIM-L and TIM-Cold could not be clearly differentiated by Western blot.
Synergism between SP and low temperature is key to trigger appropriate seasonal response in D. melanogaster. Here we found that low temperature has an enhancing effect on eya amplitude at both transcriptional and protein levels. This agrees with the transcriptomic analysis of a recent publication that reveals an up-regulation of eya in flies exposed to short photoperiod (10 L:14D) at 11 °C for 3 wk when compared to reproductively active condition (25 °C, 12 L:12D) (65). In light of the recent evidence on tim thermosensitive alternative splicing (37, 38, 66), we were able to confirm the induction of the tim-sc mRNA and protein at 10 °C. Consistent with our model, our results support the prevalent role of the cold-induced TIM-SC isoforms in stabilizing EYA under low temperature and suggest that TIM thermosensitive splicing mechanism plays a role in regulating seasonal physiology. The ability of TIM-SC to stabilize EYA appears to be due to its increased light insensitivity and affinity to EYA as compared to canonical TIM isoforms.
The model of TIM and EYA collaboration in PPTM is strongly supported by the effect of tim genetic manipulation on reproductive dormancy. Indeed, tim0 flies in SP have significantly larger ovaries compared to control individuals, whereas flies overexpressing tim undergo reproductive dormancy even in LP. All together, these results suggest that the effect of light on EYA protein stability is mediated through TIM. We should point out that our results are contrary to previous findings (34) showing that tim0 flies exhibit higher incidence of reproductive dormancy than WT flies (Canton-S). We reasoned that this apparent disparity could be due to differences in experimental conditions used to assess reproductive dormancy, genetic backgrounds of flies, or phenotypic changes due to selection in laboratory environment (see discussion below on natural tim alleles). If TIM indeed stabilizes EYA to promote reproductive dormancy, an obvious question would be regarding the nature of the cell types in which this interaction takes place. We attempted to address this question and showed that TIM-EYA interactions primarily take place in the optic lobes, which have been shown to transmit photic signals to the circadian clock neuronal network and subsequently to the PI. The fact that we did not observe TIM−EYA colocalization in the PI suggests that the mechanisms by which eya regulates seasonal physiology may differ in these two cell types.
Finally, the involvement of TIM in reproductive dormancy is supported by the findings that natural alleles of the tim loci first uncovered in Europe, ls-tim and s-tim, significantly affect the incidence of reproductive dormancy (34, 67). The derived allele ls-tim produces the longer TIM-L1421 isoform and the slightly shorter TIM-S1398 isoform, while s-tim allele only produces the latter (35). Interaction between CRY and TIM-L is weaker than with TIM-S, resulting in reduced CRY-dependent TIM degradation (35, 68) and enhanced reproductive arrest in ls-tim females (34). This is in agreement with our findings that TIM stabilizes EYA and promotes reproductive dormancy. All strains used in this study were therefore genotyped in order to identify the tim allele present in their genome and rule out tim allelic effect (SI Appendix, Table S1). With the exception of tim-overexpressing flies (w; p{tim(WT)-3XFLAG}), no correlation was found between tim allelic variation and reproductive dormancy in other strains. This indicates that the effect of eya genetic manipulation and tim0 on reproductive dormancy could not be attributed to differences in genetic background for the tim locus in those flies.
In summary, we propose that the role of EYA in seasonal adaptation in flies could rely on a combination of both internal and external coincidence through its interaction with TIM where short photoperiod restricts EYA peak to nighttime. The reliance on cold temperature by D. melanogaster to induce reproductive dormancy, a feature not necessarily shared by other insects, can be attributed to the cold induction of eya mRNA as well as EYA protein via TIM leading to increase in EYA above a specific threshold. This study provides insights to further decipher the complex mechanisms by which the circadian clock or clock-regulated proteins could inform the photoperiodic timer and allow organisms to anticipate environmental changes as seasons progress.
Material and Methods
Detailed materials and methods are provided in SI Appendix, SI Materials and Methods.
Fly Stocks.
UAS/Gal4 lines used for immunocytochemistry (ICC) and eya genetic manipulation as well as generation of tim overexpressor are described in SI Appendix.
Reproductive Dormancy Assay.
Reproductive dormancy assays are described in SI Appendix.
Whole-Mount Brain ICC.
Fly entrainment conditions and ICC procedures are described in SI Appendix.
Western Blotting and Antibodies.
Protein extraction from fly heads and Western blotting procedures were performed as described in ref. 69. Antibodies and dilutions are described in SI Appendix.
Analysis of Circadian/Daily Gene Expression by Real-Time PCR.
RNA extraction was performed as previously described (68). Primers used and real-time PCR procedures are described in SI Appendix.
Data Availability Statement.
All relevant data are within the manuscript and its SI Appendix.
Supplementary Material
Acknowledgments
We thank Patrick Emery for providing p{tim(WT)-luc} transgene, Michael Young and Deniz Top for providing α-TIM generated from N-terminal antigen, and Heinrich Jasper for providing dilp2-GS driver. We thank Takato Imaizumi, Greg Loeb, Vaughn Walton, Rufus Isaacs, Anna Wallingford, Ashfaq Ahmad Sial, and Frank Zalom for thoughtful discussion. We thank the Bloomington Drosophila Stock Center and Vienna Drosophila Resource Center for providing fly stocks, and Developmental Studies Hybridoma Bank for providing EYA antibodies. Research in the laboratory of J.C.C. is supported by NIH Grants R01 GM102225 and R01 DK124068, National Science Foundation Integrative Organismal Systems Award 1456297, and US Department of Agriculture National Institute of Food and Agriculture Grant 2015-51181-24252.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2004262117/-/DCSupplemental.
References
- 1.Emerson K. J., Bradshaw W. E., Holzapfel C. M., Complications of complexity: Integrating environmental, genetic and hormonal control of insect diapause. Trends Genet. 25, 217–225 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Nylin S., Induction of diapause and seasonal morphs in butterflies and other insects: Knowns, unknowns and the challenge of integration. Physiol. Entomol. 38, 96–104 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonini N. M., Leiserson W. M., Benzer S., The eyes absent gene: Genetic control of cell survival and differentiation in the developing Drosophila eye. Cell 72, 379–395 (1993). [DOI] [PubMed] [Google Scholar]
- 4.Jin M., Mardon G., Distinct biochemical activities of eyes absent during Drosophila eye development. Sci. Rep. 6, 23228 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duncan M. K. et al., Eyes absent: A gene family found in several metazoan phyla. Mamm. Genome 8, 479–485 (1997). [DOI] [PubMed] [Google Scholar]
- 6.Tootle T. L. et al., The transcription factor Eyes absent is a protein tyrosine phosphatase. Nature 426, 299–302 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Jemc J., Rebay I., The eyes absent family of phosphotyrosine phosphatases: Properties and roles in developmental regulation of transcription. Annu. Rev. Biochem. 76, 513–538 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Tadjuidje E., Hegde R. S., The Eyes Absent proteins in development and disease. Cell. Mol. Life Sci. 70, 1897–1913 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rebay I., Multiple functions of the eya phosphotyrosine phosphatase. Mol. Cell. Biol. 36, 668–677 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang L. et al., Eya3 partners with PP2A to induce c-Myc stabilization and tumor progression. Nat. Commun. 9, 1047 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dardente H. et al., A molecular switch for photoperiod responsiveness in mammals. Curr. Biol. 20, 2193–2198 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Masumoto K. H. et al., Acute induction of Eya3 by late-night light stimulation triggers TSHβ expression in photoperiodism. Curr. Biol. 20, 2199–2206 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Hut R. A., Photoperiodism: Shall EYA compare thee to a summer’s day? Curr. Biol. 21, R22–R25 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Wood S., Loudon A., Clocks for all seasons: Unwinding the roles and mechanisms of circadian and interval timers in the hypothalamus and pituitary. J. Endocrinol. 222, R39–R59 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wood S. H. et al., Binary switching of calendar cells in the pituitary defines the phase of the circannual cycle in mammals. Curr. Biol. 25, 2651–2662 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nelson R. J., Denlinger D. L., Somers D. E., “Photoperiodism: The biological calendar” in Photoperiodism: the Biological Calendar, (Oxford University Press, 2010). [Google Scholar]
- 17.Saunders D. S., Gilbert L. I., Regulation of ovarian diapause in Drosophila melanogaster by photoperiod and moderately low-temperature. J. Insect Physiol. 36, 195–200 (1990). [Google Scholar]
- 18.Anduaga A. M., Nagy D., Costa R., Kyriacou C. P., Diapause in Drosophila melanogaster—Photoperiodicity, cold tolerance and metabolites. J. Insect Physiol. 105, 46–53 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Nagy D. et al., A semi-natural approach for studying seasonal diapause in Drosophila melanogaster reveals robust photoperiodicity. J. Biol. Rhythms 33, 117–125 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Rayapureddi J. P. et al., Eyes absent represents a class of protein tyrosine phosphatases. Nature 426, 295–298 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Bachleitner W., Kempinger L., Wülbeck C., Rieger D., Helfrich-Förster C., Moonlight shifts the endogenous clock of Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 104, 3538–3543 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osterwalder T., Yoon K. S., White B. H., Keshishian H., A conditional tissue-specific transgene expression system using inducible GAL4. Proc. Natl. Acad. Sci. U.S.A. 98, 12596–12601 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sim C., Denlinger D. L., A shut-down in expression of an insulin-like peptide, ILP-1, halts ovarian maturation during the overwintering diapause of the mosquito Culex pipiens. Insect Mol. Biol. 18, 325–332 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schiesari L., Kyriacou C. P., Costa R., The hormonal and circadian basis for insect photoperiodic timing. FEBS Lett. 585, 1450–1460 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Sim C., Denlinger D. L., Insulin signaling and the regulation of insect diapause. Front. Physiol. 4, 189 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schiesari L., Andreatta G., Kyriacou C. P., O’Connor M. B., Costa R., The insulin-like proteins dILPs-2/5 determine diapause inducibility in Drosophila. PLoS One 11, e0163680 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y., Liao S., Veenstra J. A., Nässel D. R., Drosophila insulin-like peptide 1 (DILP1) is transiently expressed during non-feeding stages and reproductive dormancy. Sci. Rep. 6, 26620 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karpac J., Hull-Thompson J., Falleur M., Jasper H., JNK signaling in insulin-producing cells is required for adaptive responses to stress in Drosophila. Aging Cell 8, 288–295 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roman G., Davis R. L., Conditional expression of UAS-transgenes in the adult eye with a new gene-switch vector system. Genesis 34, 127–131 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Dardente H., Hazlerigg D. G., Ebling F. J., Thyroid hormone and seasonal rhythmicity. Front. Endocrinol. 5, 19 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suri V., Qian Z., Hall J. C., Rosbash M., Evidence that the TIM light response is relevant to light-induced phase shifts in Drosophila melanogaster. Neuron 21, 225–234 (1998). [DOI] [PubMed] [Google Scholar]
- 32.Myers M. P., Wager-Smith K., Rothenfluh-Hilfiker A., Young M. W., Light-induced degradation of TIMELESS and entrainment of the Drosophila circadian clock. Science 271, 1736–1740 (1996). [DOI] [PubMed] [Google Scholar]
- 33.Yang Z., Emerson M., Su H. S., Sehgal A., Response of the timeless protein to light correlates with behavioral entrainment and suggests a nonvisual pathway for circadian photoreception. Neuron 21, 215–223 (1998). [DOI] [PubMed] [Google Scholar]
- 34.Tauber E. et al., Natural selection favors a newly derived timeless allele in Drosophila melanogaster. Science 316, 1895–1898 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Sandrelli F. et al., A molecular basis for natural selection at the timeless locus in Drosophila melanogaster. Science 316, 1898–1900 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Majercak J., Sidote D., Hardin P. E., Edery I., How a circadian clock adapts to seasonal decreases in temperature and day length. Neuron 24, 219–230 (1999). [DOI] [PubMed] [Google Scholar]
- 37.Martin Anduaga A. et al., Thermosensitive alternative splicing senses and mediates temperature adaptation in Drosophila. eLife 8, e44642 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foley L. E. et al., Drosophila PSI controls circadian period and the phase of circadian behavior under temperature cycle via tim splicing. eLife 8, e50063 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bai J., Montell D., Eyes absent, a key repressor of polar cell fate during Drosophila oogenesis. Development 129, 5377–5388 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Nässel D. R., Substrates for neuronal cotransmission with neuropeptides and small molecule neurotransmitters in Drosophila. Front. Cell. Neurosci. 12, 83 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huybrechts J., De Loof A., Schoofs L., Diapausing Colorado potato beetles are devoid of short neuropeptide F I and II. Biochem. Biophys. Res. Commun. 317, 909–916 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Shimokawa K., Numata H., Shiga S., Neurons important for the photoperiodic control of diapause in the bean bug, Riptortus pedestris. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 194, 751–762 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Kubrak O. I., Kučerová L., Theopold U., Nässel D. R., The sleeping beauty: How reproductive diapause affects hormone signaling, metabolism, immune response and somatic maintenance in Drosophila melanogaster. PLoS One 9, e113051 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rieger D., Stanewsky R., Helfrich-Förster C., Cryptochrome, compound eyes, Hofbauer-Buchner eyelets, and ocelli play different roles in the entrainment and masking pathway of the locomotor activity rhythm in the fruit fly Drosophila melanogaster. J. Biol. Rhythms 18, 377–391 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Yoshii T., Hermann-Luibl C., Helfrich-Förster C., Circadian light-input pathways in Drosophila. Commun. Integr. Biol. 9, e1102805 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saunders D. S., Henrich V. C., Gilbert L. I., Induction of diapause in Drosophila melanogaster: Photoperiodic regulation and the impact of arrhythmic clock mutations on time measurement. Proc. Natl. Acad. Sci. U.S.A. 86, 3748–3752 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tatar M., Yin C., Slow aging during insect reproductive diapause: Why butterflies, grasshoppers and flies are like worms. Exp. Gerontol. 36, 723–738 (2001). [DOI] [PubMed] [Google Scholar]
- 48.Saunders D. S., Insect photoperiodism: Seeing the light. Physiol. Entomol. 37, 207–218 (2012). [Google Scholar]
- 49.Iiams S. E., Lugena A. B., Zhang Y., Hayden A. N., Merlin C., Photoperiodic and clock regulation of the vitamin A pathway in the brain mediates seasonal responsiveness in the monarch butterfly. Proc. Natl. Acad. Sci. U.S.A. 116, 25214–25221 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hazlerigg D., The evolutionary physiology of photoperiodism in vertebrates. Prog. Brain Res. 199, 413–422 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Shim J. S., Kubota A., Imaizumi T., Circadian clock and photoperiodic flowering in Arabidopsis: CONSTANS is a hub for signal integration. Plant Physiol. 173, 5–15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johnston J. D. et al., Multiple effects of melatonin on rhythmic clock gene expression in the mammalian pars tuberalis. Endocrinology 147, 959–965 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Mohamed A. A. et al., N-acetyltransferase (nat) is a critical conjunct of photoperiodism between the circadian system and endocrine axis in Antheraea pernyi. PLoS One 9, e92680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barberà M. et al., Melatonin in the seasonal response of the aphid Acyrthosiphon pisum. Insect Sci. 27, 224–238 (2020). [DOI] [PubMed] [Google Scholar]
- 55.Bertolini E. et al., Life at high latitudes does not require circadian behavioral rhythmicity under constant darkness. Curr. Biol. 29, 3928–3936.e3 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Lear B. C. et al., A G protein-coupled receptor, groom-of-PDF, is required for PDF neuron action in circadian behavior. Neuron 48, 221–227 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Meuti M. E., Denlinger D. L., Evolutionary links between circadian clocks and photoperiodic diapause in insects. Integr. Comp. Biol. 53, 131–143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ikeno T., Numata H., Goto S. G., Shiga S., Involvement of the brain region containing pigment-dispersing factor-immunoreactive neurons in the photoperiodic response of the bean bug, Riptortus pedestris. J. Exp. Biol. 217, 453–462 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Hand S. C., Denlinger D. L., Podrabsky J. E., Roy R., Mechanisms of animal diapause: Recent developments from nematodes, crustaceans, insects, and fish. Am. J. Physiol. Regul. Integr. Comp. Physiol. 310, R1193–R1211 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Menegazzi P. et al., Adaptation of circadian neuronal network to photoperiod in high-latitude European Drosophilids. Curr. Biol. 27, 833–839 (2017). [DOI] [PubMed] [Google Scholar]
- 61.King A. N., Sehgal A., Molecular and circuit mechanisms mediating circadian clock output in the Drosophila brain. Eur. J. Neurosci. 51, 268–281 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kozak G. M. et al., Genomic basis of circannual rhythm in the European corn borer moth. Curr. Biol. 29, 3501–3509.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Ojima N., Hara Y., Ito H., Yamamoto D., Genetic dissection of stress-induced reproductive arrest in Drosophila melanogaster females. PLoS Genet. 14, e1007434 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nagy D. et al., Peptidergic signaling from clock neurons regulates reproductive dormancy in Drosophila melanogaster. PLoS Genet. 15, e1008158 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kučerová L. et al., Slowed aging during reproductive dormancy is reflected in genome-wide transcriptome changes in Drosophila melanogaster. BMC Genomics 17, 50 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shakhmantsir I., Nayak S., Grant G. R., Sehgal A., Spliceosome factors target timeless (tim) mRNA to control clock protein accumulation and circadian behavior in Drosophila. eLife 7, e39821 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zonato V., Vanin S., Costa R., Tauber E., Kyriacou C. P., Inverse European latitudinal cline at the timeless Locus of Drosophila melanogaster reveals selection on a clock gene: Population genetics of ls-tim. J. Biol. Rhythms 33, 15–23 (2018). [DOI] [PubMed] [Google Scholar]
- 68.Peschel N., Chen K. F., Szabo G., Stanewsky R., Light-dependent interactions between the Drosophila circadian clock factors cryptochrome, jetlag, and timeless. Curr. Biol. 19, 241–247 (2009). [DOI] [PubMed] [Google Scholar]
- 69.Chiu J. C., Vanselow J. T., Kramer A., Edery I., The phospho-occupancy of an atypical SLIMB-binding site on PERIOD that is phosphorylated by DOUBLETIME controls the pace of the clock. Genes Dev. 22, 1758–1772 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are within the manuscript and its SI Appendix.