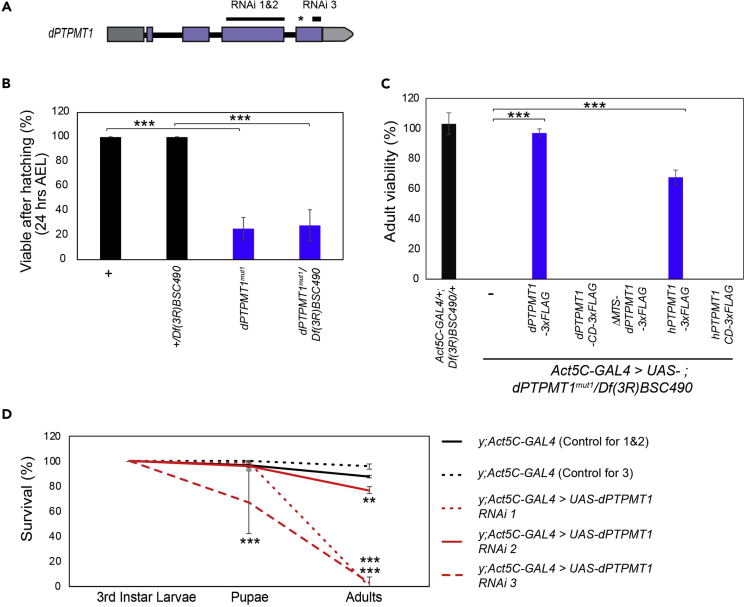
Figure 1.
dPTPMT1 Is an Essential Gene in Drosophila
(A) Regions in dPTPMT1 targeted by the three RNAi lines. Thin black bars indicate regions targeted by RNAi, dark gray bar is the 5′ UTR, light gray bar is the 3′ UTR, purple bars are exons, and thick black bars are introns. Asterisk indicates the location of the deletion in dPTPMT1mut1.
(B) Viability of dPTPMT1mut1 first instar larvae at 24 h AEL. n = 4–5 replicates, 56–285 for each repeat. Data are represented as mean ± SEM and ∗∗∗p < 0.001 by chi-square test.
(C) Adult viability of dPTPMT1mut1 mutants. Genotypic frequency is calculated for adults and then normalized to expected genotypic frequency. n = 3–4 replicates; 0- to 2-day-old adults. Data are represented as mean ± SEM and ∗∗∗p < 0.001 by chi-square test.
(D) Percentage of third instar males with ubiquitous dPTPMT1 RNAi surviving to pupation and eclosion. Larvae are on a yellow mutant background (hereafter indicated by “y”). Statistical significance is given relative to Act5C-GAL4 crossed to the respective RNAi background controls at the same developmental stage. n = 3 replicates, 25–26 for each repeat for RNAi 1-2 and controls. n = 3 replicates, 12–44 for each repeat for RNAi 3 and control.
Data are represented as mean ± SEM and ∗∗p < 0.01, ∗∗∗p < 0.001 by chi-square test. See also Figure S1.