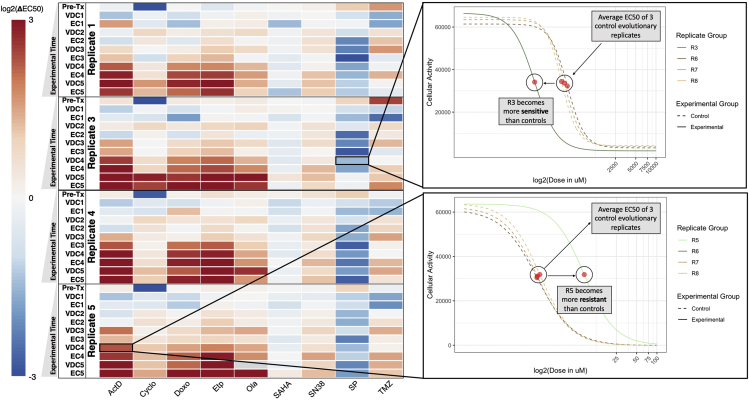
Figure 2.
Temporal Collateral Sensitivity Map Representing EC50 Changes to a Panel of Drugs as the A673 Cell Line Develops Resistance to Standard Treatment
Left: A heatmap representing how the EC50 to a panel of nine drugs changes in four A673 cell line evolutionary replicates as they are exposed to the VDC/EC drug combinations over time. Color represents the log2 fold change of EC50 to a drug (columns) for a replicate at a given evolutionary time point (rows) compared with the average EC50 of the three control evolutionary replicates at the corresponding time point. Values above log2(3) or below log2(-3) are represented by log2(3) and log2(-3), respectively. Time points are denoted as the drug combination that a given replicate has recently recovered from. For example, the data representing dose-response models after the first application of the VDC drug combination would be labeled with VDC1. Of note, the EC50 of olaparib in Replicate 5 at the VDC5 time point is indeterminate owing to a poorly fit dose-response model. This value in the heatmap is denoted as gray, but Figure S1 remains uncensored. Right: Top, a plot of the dose-response curves for Replicate 3 and all control replicates (Replicates 6, 7, 8) in response to SP-2509 (SP) at the VDC4 time point. Bottom, a plot of the dose-response curve for Replicate 5 and all control replicates in response to dactinomycin at the VDC4 time point. Cellular activity is measured by enzymatic conversion of alamarBlue, normalized to background florescence. Estimated EC50 for each replicate is denoted with a red circle. These two dose-response plots demonstrate how the heatmap (left) values were calculated, where the control EC50 values are averaged, and the heatmap values represent the log2 fold change between a given replicate and this mean EC50 value.