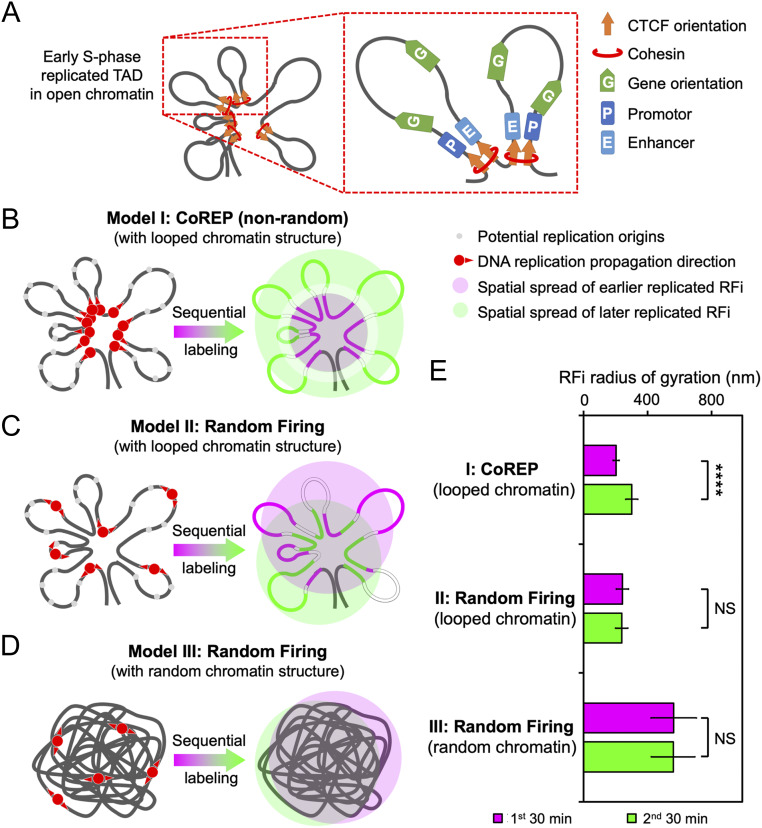
Fig. 6.
CoREP model for nonrandom replication activation and propagation. (A) Schematic diagram of an early S-phase TAD, organized into looped structures by CTCF and cohesin located at the base; each loop contains promotors, enhancers, and active genes. (B–D) Three potential models for the spatiotemporal propagation of DNA replication in early S-phase: CoREP model (B), in which replication is preferentially activated on CTCF-organized looped chromatin structures; random firing model with looped chromatin structures (C); and random firing model with random chromatin structures (D). (E) Comparison of simulated radii of gyration of RFi produced in two consecutive 30-min labeling windows, using the three models described in B–D. Error bars denote mean ± SD. P values are determined by two-tailed Student’s t test; ****P < 0.0001; NS: not significant. n = 500 chromatin configurations for E.