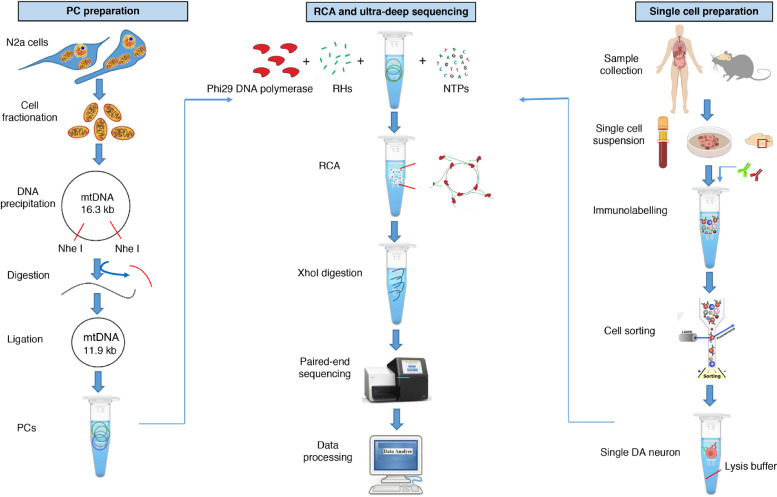
Fig. 1.
Overview of MitoSV-seq method. Samples were collected from mouse brain, cancer cell lines, and human buffy coats. Single cells were isolated by FACS using cell-specific markers. PC preparation is optional, and although it is not necessary, it is highly recommended for mtDNA sequencing. PCs and single-cell lysates were amplified by RCA. High-molecular-weight concatemers were digested using XhoI. Tagmentation and indexing were performed using the Nextera XT DNA Library Preparation Kit. Libraries were pooled and run for paired-end 300-cycle high-mode sequencing on a NextSeq 500. NGS data were analysed using standard DNA-seq methods. Pindel [58] was applied for SV calling. N2a, Neuro-2a; mtDNA, mitochondrial DNA; PC, positive control; RCA, rolling circle amplification; RHs, random hexamers; dNTP, deoxyribonucleotide triphosphate; DA, dopaminergic.