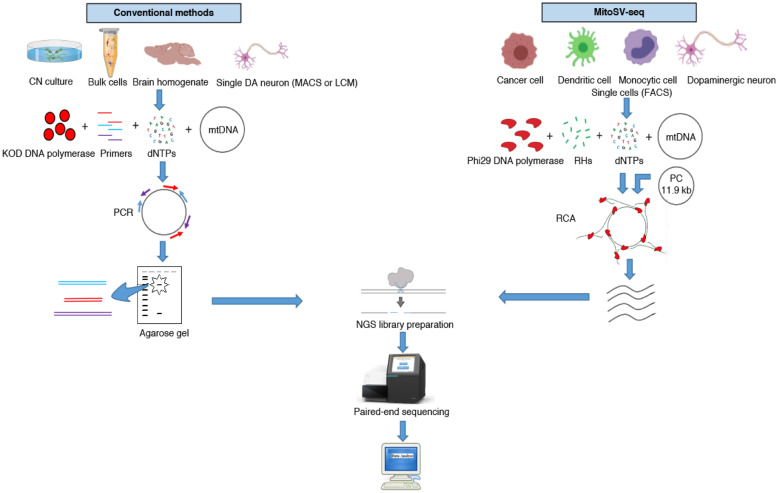
Fig. 2.
Step-by-step schematic between MitoSV-seq and conventional methods. Conventional methods have focused primarily on brain homogenate, bulk cortical neurons (CNs), or single DA neurons isolated by MACS or LCM, followed by PCR amplification of mtDNA using site-specific primers designed mainly for the D-loop or major arc. Amplified PCR fragments are usually purified from agarose gels and processed for NGS library preparation and sequencing. MitoSV-seq benefits from FACS in characterising and isolating single cells from a heterogeneous pool of cells using cell-specific markers. Single cells are sorted and used directly for the amplification step. Concatemers from circular mtDNA are generated using RHs and phi29 DNA polymerase in a rolling circle manner. Addition of our custom PC allows to us to first set up the analytical pipeline to find a known PC deletion. High-molecular-weight concatemers are digested and used for NGS library preparation and ultra-deep sequencing. MACS, magnetic cell separation; LCM, laser capture microdissection; PCR; polymerase chain reaction; FACS; fluorescence-activated cell sorting; RHs, random hexamers; RCA, rolling circle amplification; PC, positive control.