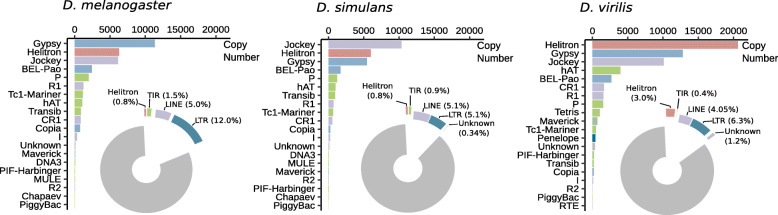
Fig. 2.
TE contents in D. melanogaster, D. simulans and D. virilis (from left to right). Barplots represent TE copy numbers for the top 20 TE superfamilies. Piecharts illustrate genomic sequence occupancy of each TE order (in percentages of the assemblies). These results were obtained using the D. melanogaster reference genome assembly (r6.29), and recently produced long-reads assemblies of D. simulans and D. virilis [99]. RepeatMasker was used to recover TE fragments and TE genomic sequence occupancy (RepeatMasker v1.332, -nolow, -norna, -species drosophila; Repbase-derived RepeatMasker libraries 20181026 [100],). TE fragments were assembled into TE copies using OneCodeToFindThemAll [101].