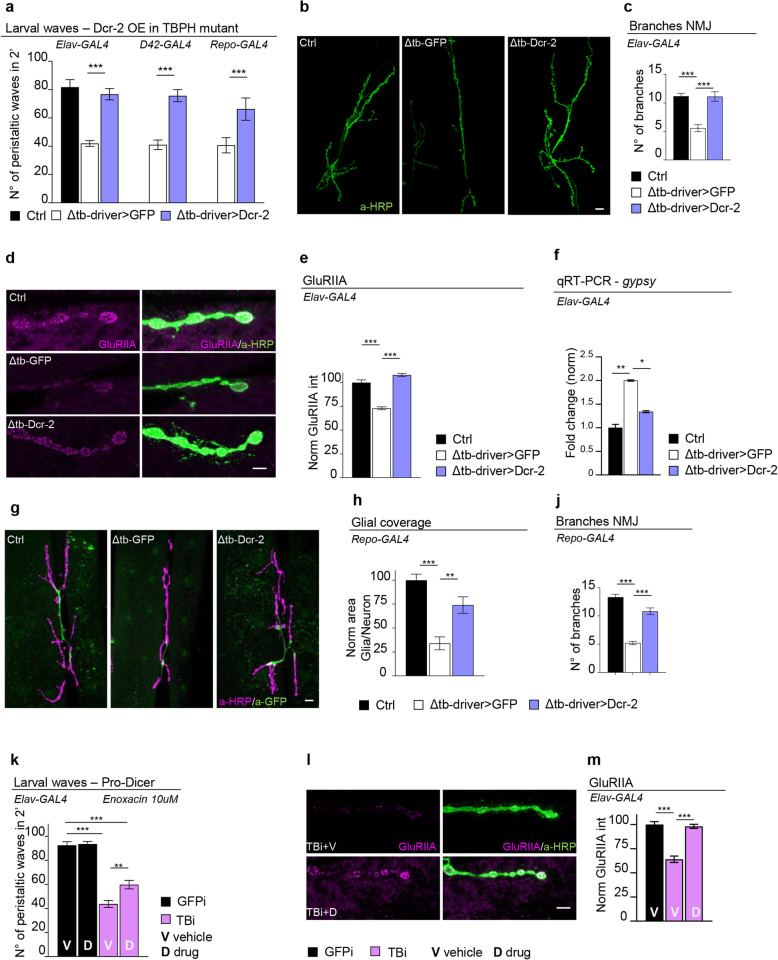
Fig. 4.
Genetic rescue of Dcr-2 expression recovers TBPH mutant pathological phenotypes. a Number of peristaltic waves of Ctrl (w1118), Δtb-driver>GFP (tbphΔ23/tbphΔ23;driver-GAL4/UAS-GFP), and Δtb-driver>Dcr-2 (UAS-Dcr-2/+;tbphΔ23/tbphΔ23;driver-GAL4/+). elav-GAL4, D42-GAL4, and repo-GAL4 were used as reported on the figure. n = 20. b Confocal images of third instar NMJ terminals in muscle 6/7 second segment stained with anti-HRP (in green) in Ctrl, Δtb-driver>GFP, and Δtb-driver>Dcr-2, using elav-GAL4. c Quantification of branch number. n = 15. d Confocal images of third instar NMJ terminals in muscle 6/7 second segment stained with anti-HRP (in green) and anti-GluRIIA (in magenta) in Ctrl, Δtb-driver>GFP, and Δtb-driver>Dcr-2, using elav-GAL4. e Quantification of GluRIIA intensity. n > 200 boutons. f Real-time PCR of gypsy transcript levels normalized on Rpl11 (housekeeping) in Ctrl, Δtb-driver>GFP, and Δtb-driver>Dcr-2, using elav-GAL4. n = 2, error bars SEM. g Confocal images of third instar NMJ terminals in muscle 6/7 second segment stained with anti-HRP (in magenta) for presynaptic membrane and anti-GFP (in green) for peripheral glia in Ctrl (+/+;repo-GAL4,UAS-GFP/+), Δtb-GFP (tbphΔ23/tbphΔ23;repo-GAL4,UAS-GFP/+), and Δtb-Dcr-2 (UAS-Dcr-2/+;tbphΔ23/tbphΔ23;repo-GAL4,UAS-GFP/+). h Quantification of glial area. n = 10 larvae. j Quantification of branch number. n = 10. k Number of peristaltic waves of GFPi (Dcr-2/+; tbphΔ23,elav-GAL4/+;UAS-GFP-IR/+) and TBi (Dcr-2/+; tbphΔ23,elav-GAL4/+;UAS-TBPH-IR/+) fed with 10 μM enoxacin (D) compared to vehicle only (V). n = 20. l Confocal images of third instar NMJ terminals in muscle 6/7 second segment stained with anti-HRP (in green) and anti-GluRIIA (in magenta) in GFPi and TBi. m Quantification of GluRIIA intensity. n > 200 boutons. *p < 0.05, **p < 0.01, ***p < 0.001 calculated by one-way ANOVA, error bars SEM. Scale bar 10 μm (b, g) and 5 μm (d, l)