Fig. 1.
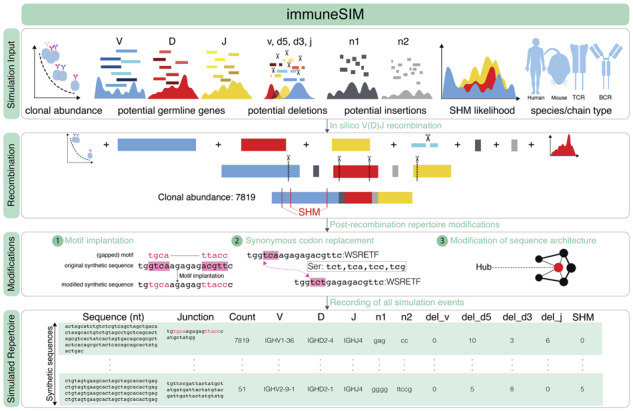
immuneSIM simulates fully (single, paired) annotated tunable immune repertoires that are either highly similar (native-like) or deviating (aberrant, see main text for definition) from experimental immune repertoires. All major immune repertoire features, such as clonal abundance, germline genes, deletions and insertions and somatic hypermutation, are tunable. Post in silico recombination, the immuneSIM-generated immune receptor repertoires may be further modified by (i) implantation of motifs, (ii) codon replacement and (iii) change of sequence similarity architecture
