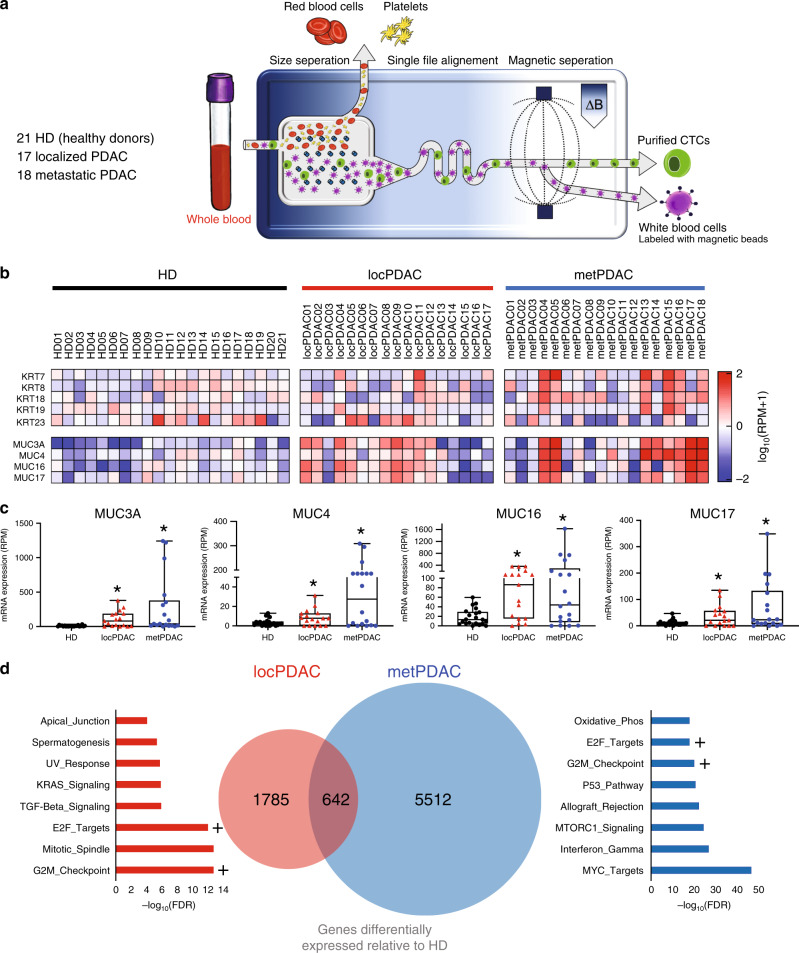
Fig. 1. Transcriptional profiling of CTCs purified from the blood of patients with pancreatic cancer reveals substantial heterogeneity.
a Schematic of circulating tumor cell purification, via removal of erythrocytes and platelets followed by efficient magnetophoretic depletion of labeled leukocytes, utilizing the CTC-iChip. b Heatmap of log10(RPM+1) mRNA expression of keratin and mucin genes in CTCs purified from the blood of patients with localized PDAC (locPDAC, n = 17), and metastatic PDAC (metPDAC, n = 18) compared with healthy donor (HD, n = 21) controls. c Box plots showing expression of 4 mucin genes detected in cells released from localized PDAC and metastatic PDAC. *FDR < 0.10 relative to HDs of DESeq2-normalized data; n = 21 HD, 17 locPDAC, 18 metPDAC; median and 25–75% IQR shown by box plots. d Venn diagram of genes upregulated (17 locPDAC and 18 metPDAC compared with specimens from 21 HD controls; *FDR < 0.10) in localized PDAC and metastatic PDAC CTCs; “Hallmark” gene set ontology terms for each group are shown. + Denotes common ontology terms shared between both groups.