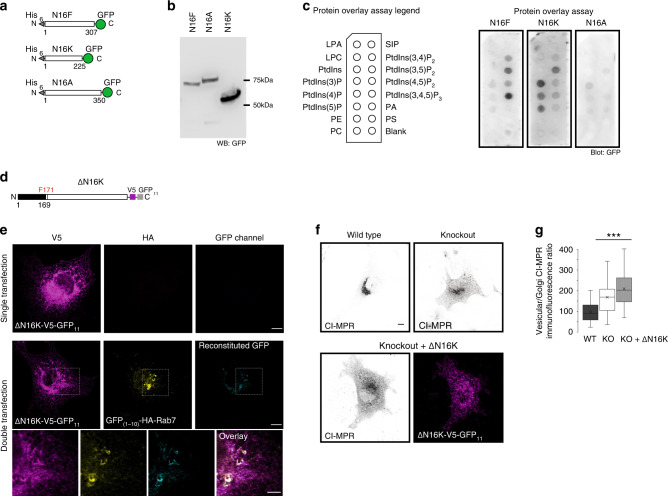
Fig. 6. TMEM16K N-terminal domain.
a Scheme of purification constructs of N-terminal cytosolic domains of TMEM16F (N16F, predicted MW 63.6 kDa), TMEM16K (N16K, predicted MW 53.4 kDa) and TMEM16A (N16A, predicted MW 67.5 kDa) b Western blot of purified N-terminal cytosolic domains revealed with anti-GFP antibody and horseradish peroxidase. c Lipid binding of predicted phosphatidylinositol binding domains was evaluated by loading 10 µg of purified proteins to the PIP-strips and visualized with blotting with anti-GFP antibody and horseradish peroxidase. Representative images of three independent blots. d Scheme of N-terminal truncation mutant of TMEM16K where 1–169 amino acids were deleted. e Split-GFP reconstitution assay to evaluate N-terminal truncation mutant for its ability to reconstitute GFP with Rab7. Cell were single or double transfected cells with ΔN –terminal TMEM16K-V5-GFP11 and GFP(1–10) tagged Rab7, Scale bar 10 µm for rows 1, 2, and 5 µm for inset in the row 3. f Representative images of the ability of ΔN16K-V5-GFP11 to rescue endosomal retrograde trafficking defect when introduced in TMEM16K KO cells as measured by CI-MPR assay described in Fig. 3a (WT and KO from Fig. 3a repeated for clarity of comparison) Scale bar 10 µm. g Quantification (same data from Fig. 3a added to this graph for clarity of comparison). Single factor ANOVA, p value = 2.03E−49 with post-test Bonferroni-corrected two sided t-test with pairwise comparison with WT (three biological replicates, n = 168 WT, 181 TMEM16K KO, p value = 1.37E−26*** and 161 TMEM16K KO + ΔN16K-V5-GFP11 cells, p value = 6.06E−47***). In the box and whiskers plot, the box includes the first quartile and the third quartile, with the central line representing the median. Whiskers represent the minimum and maximum values of data. X inside the box represents the mean of data. Source data are provided as a Source Data file.