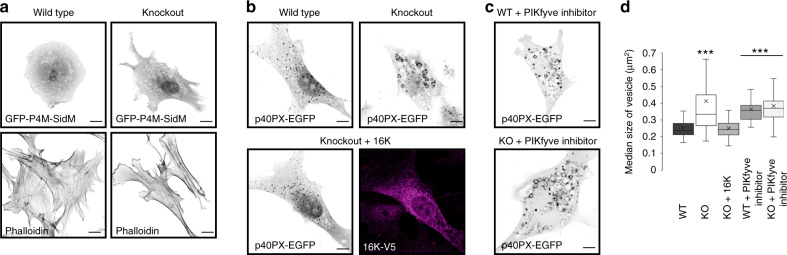
Fig. 7. Endolysosomal phosphatidylinositols.
a Confocal images of WT and TMEM16K KO cells transfected with GFP-P4M-SidM, biosensor for PtdIns4P or labeled with fluorescence conjugated phalloidin which specifically labels actin network. Representative images from two independent experiments. Scale bar 10 µm. b. Confocal images of WT, TMEM16K KO cells and KO cells with reintroduced TMEM16K cells transfected with PtdIns3P biosensor P40PX-EGFP. Scale bar 10 μm. c. Confocal images of WT and TMEM16K KO cells transfected with PtdIns3P biosensor P40PX-EGFP and treated with PIKfyve kinase inhibitor YM201636. Scale bar 10 μm. d. Quantification of the median size of the PtdIns3P positive vesicles per cell visualized with P40PX-EGFP from 3 biological replicates in the WT, TMEM16K KO and TMEM16K KO cells transfected with wild type TMEM16K cDNA, as well as WT and TMEM16K KO cell treated with YM201636. Single factor ANOVA, p value = 6.82E−63 with post-test Bonferroni-corrected two sided t-test with pairwise comparison with WT. (n = 131 WT, n = 131 KO, p value = 1.50E−18***, n = 153 KO + 16K, n = 150 WT + PIKfyve inhibitor, p value = 5.86E−42***, n = 156 KO + PIKfyve inhibitor, p value = 1.59E−41***). In the box and whiskers plot, the box includes the first quartile and the third quartile, with the central line representing the median. Whiskers represent the minimum and maximum values of data. X inside the box represents the mean of data. Source data are provided as a Source Data file.