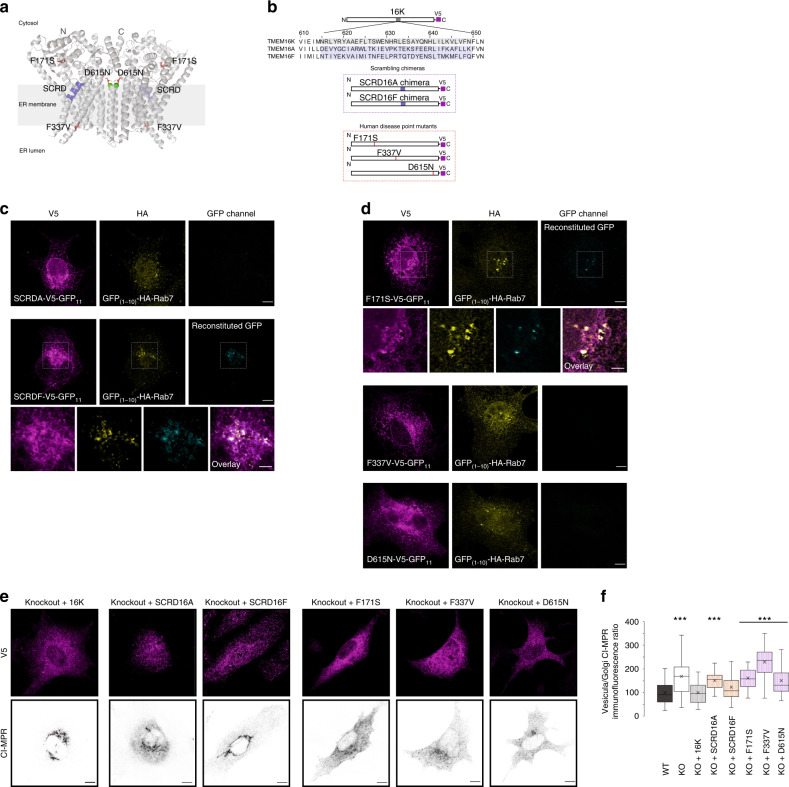
Fig. 8. TMEM16K scrambling domain chimeras and human disease variants.
a Topology of TMEM16K dimer in the endoplasmic reticulum membrane with labeled human disease point mutations and SCRD (minimal lipid scrambling domain) based on published crystal structure33 and represented in Pymol. b Scheme of the SCRD chimeras and human disease point mutation constructs c Split-GFP reconstitution assay of scrambling domain chimeras (SCRDA, SCRDF) tagged with GFP11 and GFP(1-10) tagged Rab7. Scale bar 10 µm, inset 5 µm. d Split-GFP reconstitution assay of human point mutants (F171S, F337V, D615N) tagged with GFP11 with GFP(1-10) tagged Rab7. Scale bar 10 µm, inset 5 µm. e Representative images of the ability of SCRD chimeras and human disease point mutants to rescue endosomal retrograde trafficking defect when introduced in TMEM16K KO (KO + 16K cDNA from Fig. 3a repeated for clarity of comparison) Scale bar 10 µm. f Quantification done as in Fig. 3a (same data added to this graph for clarity of comparison). Single factor ANOVA, p value = 2.1E−117 with post-test Bonferroni-corrected two sided t-test with pairwise comparison with WT, (three biological replicates, n = 168 WT, 181 TMEM16K KO, p value = 1.37E−26*** and 134 KO + 16K cells, 130 KO + 16KSCRD16A, p value = 5.05E−29***, 145 KO + 16KSCRD16F, n = 145 KO + F171S, p value = 1.98E−36***, 138 KO + F337V, p value = 1.37E−17***, 155 KO + D615N cells, p value=1.37E-29***). In the box and whiskers plot, the box includes the first quartile and the third quartile, with the central line representing the median. Whiskers represent the minimum and maximum values of data. X inside the box represents the mean of data. Source data are provided as a Source Data file.