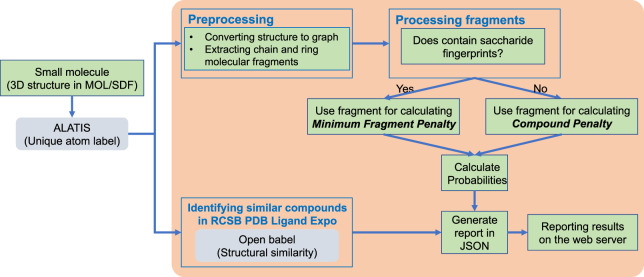
Fig. 6.
Workflow of the web server. For a given small molecule, the web server queries ALATIS to retrieve unique atom labels of the compound. The preprocessing module converts the structure file to a graph data structure, and extracts chain and ring molecular fragments. Then every fragment is analyzed to identify saccharide fingerprints. If no fingerprint found, the fragment is used in calculating a compound penalty. The molecular fragments that contain saccharide fingerprints are used in calculating the minimum fragment penalty. This penalty and the compound penalty are then used in calculating the probabilities. The web server reports the calculated probabilities and also lists every other calculated fragment penalty for the molecular fragments. In parallel, the web server uses the Open Babel package for identifying ligands with the highest structural similarities to the submitted molecule. These ligands from the RCSB PDB Ligand Expo are cross-referenced to the PDB molecular complexes. The outcome of this structural analysis reports proteins from PDB that bind to the identified ligands.