Figure 2. The Dilated Form of DnaB Interacts Tightly with bioχψτ3δδ’.
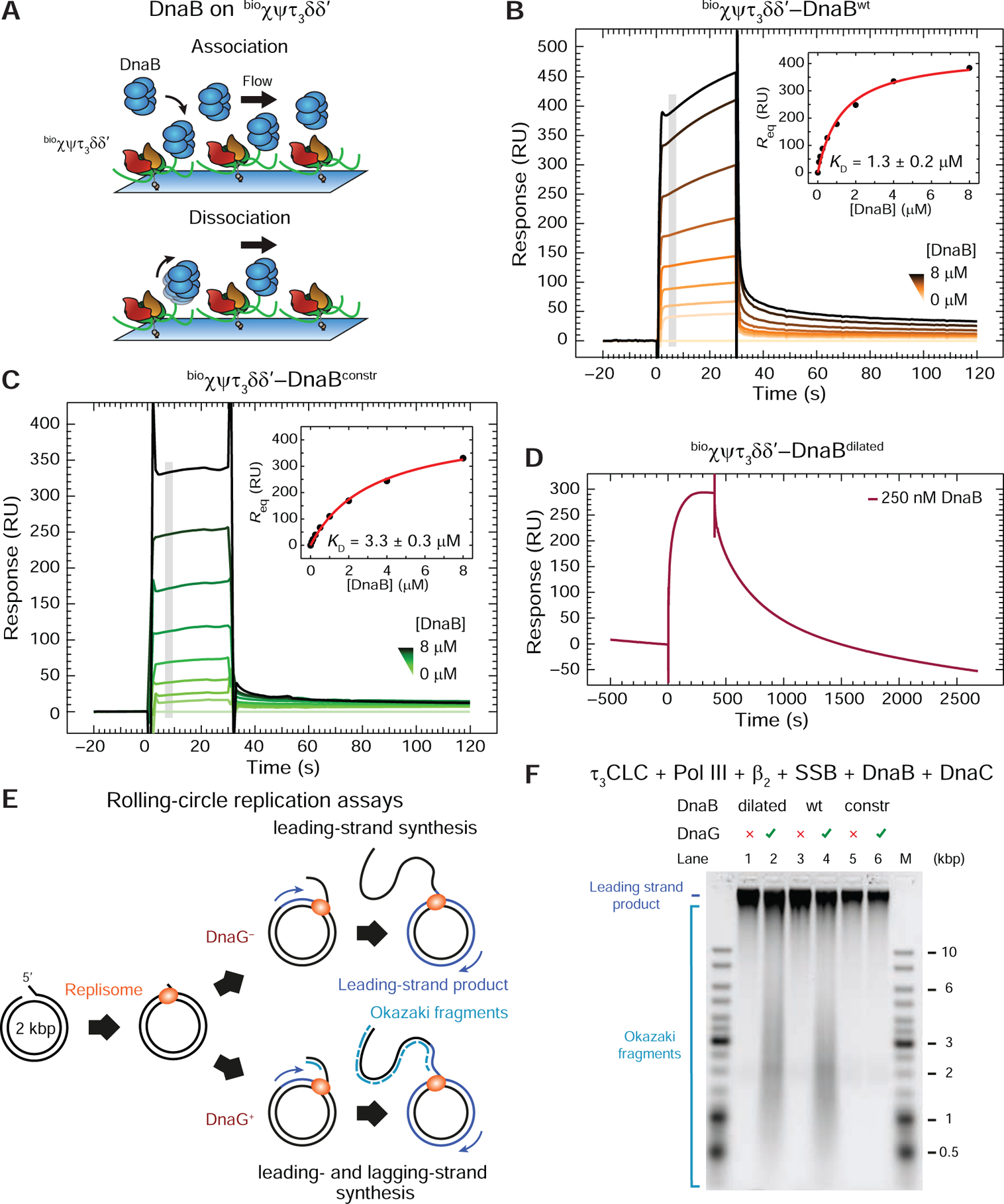
(A) Cartoon representation of the SPR experiment used to measure the binding of DnaB versions in solution to immobilized τ3CLC.
(B) and (C) SPR sensorgrams show association (30 s) and dissociation phases of serially-diluted 0.0625–8 μM DnaBwt (B) and DnaBconstr (C) on bioχψτ3δδ’, including 0 μM control. Responses at equilibrium, determined by averaging values in the gray region, were fit (insets) using a 1:1 steady state affinity (SSA) model to derive values of KD (as indicated) and Rmax = 440 ± 20 RU (B), 460 ± 20 RU (C). Errors are S.E. of the fit.
(D) DnaBdilated (250 nM) is injected for 400 s and its dissociation monitored over 2000 s.
(E) Representation of bulk replication assays with rolling-circle substrates in the absence or presence of primase to enable lagging-strand synthesis.
(F) Alkaline agarose gel showing resolved leading- (all lanes) and lagging-strand (DnaG+ lanes) products generated by replisomes in the absence of primase (odd lanes) or its presence (even lanes), using DnaBdilated, DnaBwt, and DnaBconstr (as indicated).
See also Figure S1.
