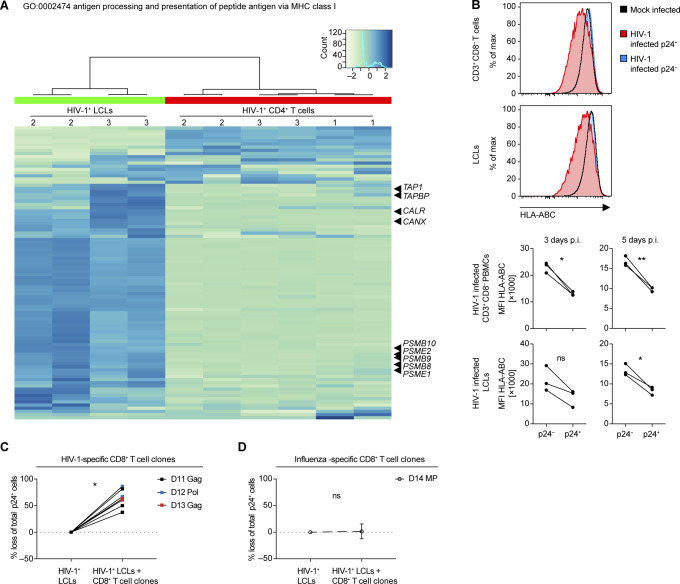
Figure S5. Effect of HIV-1 infection in lymphoblastoid cell lines (LCLs) and T cells on genes in the GO set “0002474 antigen processing and presentation of peptide antigen via MHC class I,” HLA class I surface expression and specific elimination of HIV-1 infected LCLs via HIV-1 specific CD8+ T cell clones.
(A) Heat map of relative gene expression in the gene set “GO:0002474 antigen processing and presentation of peptide antigen via MHC class I” of host transcriptome of HIV-infected LCLs and CD4+ T cells. Indicated genes: TAP1 (peptide transporter TAP1), TAPBP (Tapasin), CALR (calreticulin), CANX (calnexin), PSMB10 (Mecl-1), PSME2 (Pa28β), PSMB9 (LMP2), PSMB8 (LMP7), and PSME1 (Pa28α). (B) Representative flow cytometry plots 3 d post HIV-1 infection (top panel) and summary of median fluorescence intensity of HLA class I (pan-A/B/C) detected on the surface of mock-infected or HIV-1–infected (p24− and p24+ populations) CD3+ CD8− T cells and LCLs 3 and 5 d postinfection for three donors (bottom panel) (two-tailed unpaired t test). (C) Specific elimination of p24+ autologous or HLA-matched LCLs by three individual HIV-1–specific CD8+ T-cell clones expressed as percentage loss of total p24+ cells. (D) Specific elimination of p24+ LCLs by influenza-specific CD8+ T-cell clones (Influenza MP [GIL] - specific) expressed as percentage loss of total p24+ cells. Data from two experiments using different (matched) target cells each time. (C, D) ns P > 0.05, *P < 0.05, **P < 0.01 (Wilcoxon matched-pairs test).