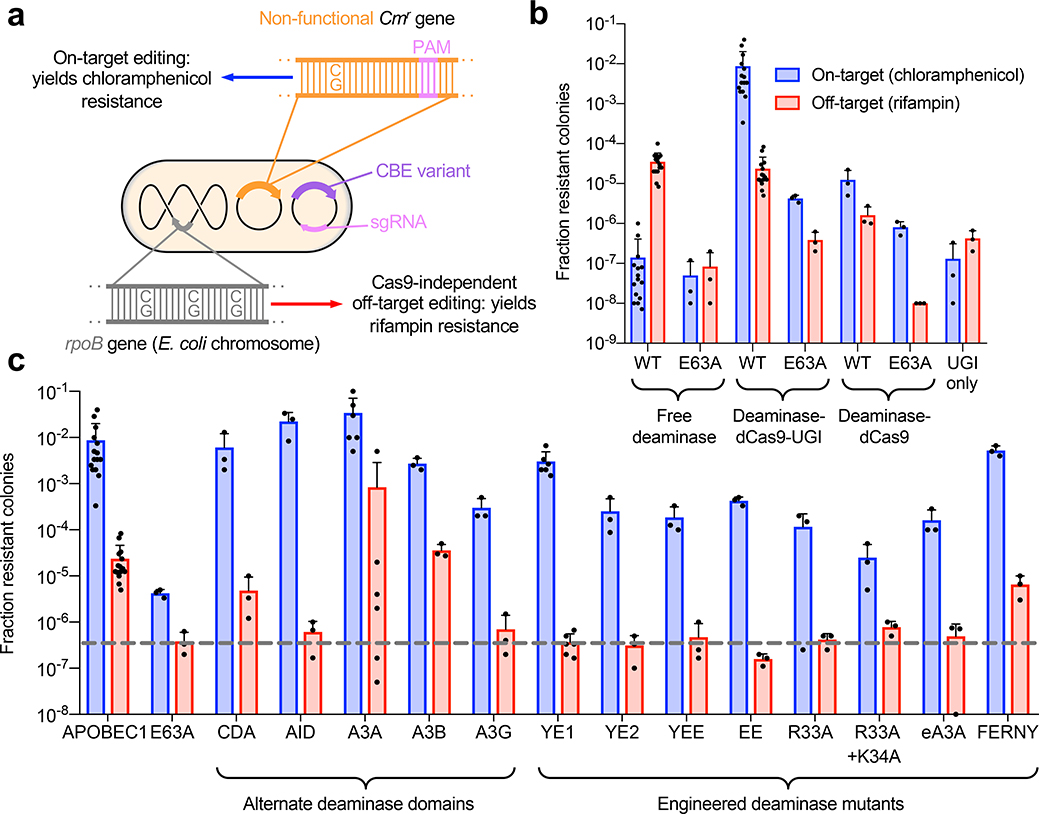
Figure 1. On-target and Cas9-independent off-target DNA editing in E. coli.
(a) Experimental design. (b) Assay validation. Fraction of resistant colonies was calculated relative to number of E. coli plated on maintenance antibiotics. Data are shown as individual data points and mean ± SEM for n=3 or n=15 bacterial colonies. E63A refers to a catalytically inactivated APOBEC1 E63A mutant. (c) Performance of CBEs that use alternative deaminases. All constructs are in the deaminase–dCas9–UGI (BE2) architecture. The grey dotted line indicates the background level of rifampin resistance of the inactive APOBEC1 E63A deaminase control. Data are shown as individual data points and mean ± SEM for n=3 or n=15 bacterial colonies.