Abstract
The penta-substituted pyridine natural products harzianopyridone and atpenins are potent inhibitors of mitochondrial complex II. We identified the pathways of these compounds from their fungal producers, and uncovered the biosynthetic steps require multiple iterative enzymes. In particular, a methyltransferase and a flavin-dependent monooxygenase are used iteratively to introduce C5 and C6 methoxy groups. The pathway unexpectedly requires the installation and removal of a N-methoxy group, which is proposed to be a directing group that tunes the reactivity of the pyridone ring, possibly through the alpha effect.
Graphical Abstract
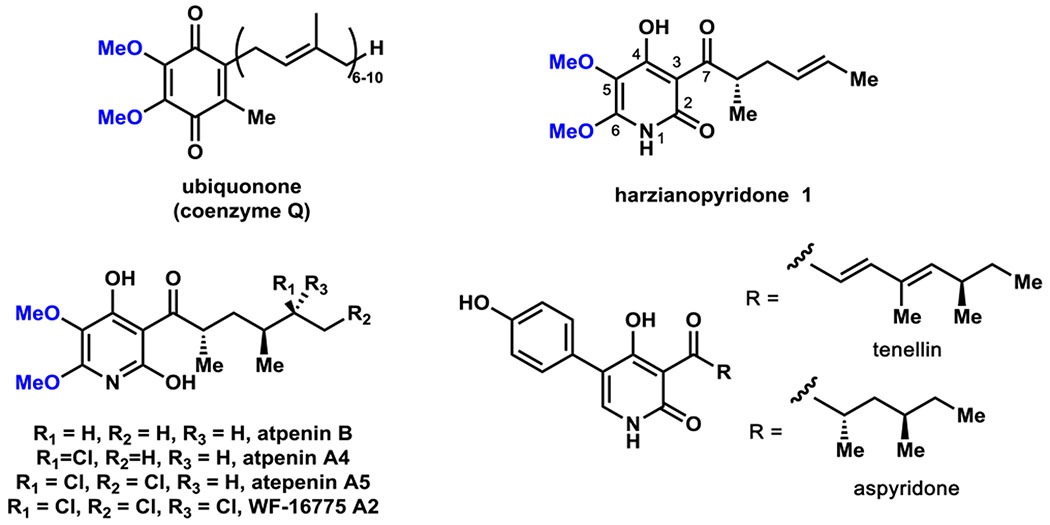
Oxidative phosphorylation (OXPHOS) is an energy generating process that takes place in the inner membrane of prokaryotic and mitochondria of eukaryotic organisms, and is an attractive target for antifungal and anticancer therapies.1 Potent natural products that selectively target one of the five complexes (I-V) in OXPHOS have been identified and have entered clinical development.2 Among them, the fungal 2-pyridones harzianopyridone 13 and atpenin A54 are nanomolar inhibitors of mammalian succinate ubiquinone oxidoreductase (mitochondrial complex II, CII) (Figure 1).5 Structures of these compounds closely match that of the electron carrier ubiquinone with their penta-substituted pyridine cores.6 The C5 and C6 methoxy groups in 1 and atpenin match to those present in ubiquinone, and are not found in any other 2-pyridone natural products. 7 Because of the exceptionally potent activities of 1 and atpenin A5, these compounds have been the subjects of numerous total synthesis efforts,8–11 as well as structure-activity-relationship studies.12,13 The synthetic strategy relies on the C3-C7 coupling between a fully functionalized organometallic pyridine nucleophile with the corresponding acyl aldehyde, both requiring lengthy synthetic steps to prepare. It is therefore interesting to compare Nature’s logic in enzymatically generating the heavily substituted pyridone compounds. However, the biosynthetic strategies to produce 1 and atpenin are not known; and no biosynthetic gene clusters (BGCs) have been reported.
Figure 1.
Structures of ubiquinone and 2-pyridone natural products.
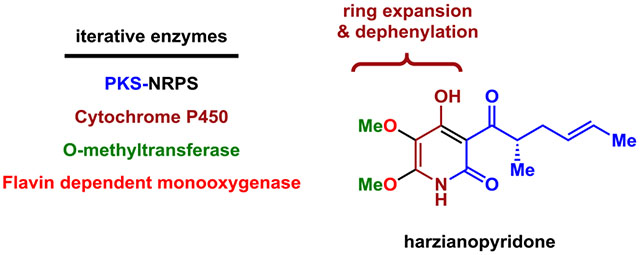
Based on the biosynthesis of other 2-pyridone natural products such as tenellin,14 (Figure 1) as well as isotope feeding studies of 1,15 both 1 and atpenin are proposed to derive from a polyketide-amino acid containing tetramic acid, which is produced by a polyketide synthase-nonribosomal peptide synthetase (PKS-NRPS). In the pathways of tenellin16 and leporin,17 a ring expansion P450 (P450RE) catalyzes a radical-mediated ring expansion to afford a 4-hydroxy-3-acyl-2-pyridone intermediate. Subsequent hydroxylations and methylations catalyzed by flavoenzymes and methyltransferases, respectively, can be proposed to install the C5- and C6-methoxy groups. Guided by this proposal, we searched for a candidate BGC of 1 from the genome of Trichoderma harzianum, a reported producer.3 We found one BGC, renamed the har BGC, to contain homologs of both PKS-NRPS (HarA) and P450RE (HarG) (Figure 2A and Table S1). Also encoded in the gene cluster are the enoylreductase (ER) (HarE) that partners with PKS-NRPS, two flavin-dependent monooxygenases (FMOs) (HarC and HarF), an additional P450 (HarD), and an O-methyltransferase (O-MT) (HarB). HarD is predicted to be a N-hydroxylase based on homology to characterized enzymes. 18 The functional predictions of the genes in the cluster are therefore consistent with the structural features in 1, with the exception of the putative N-hydroxylase HarD.
Figure 2.
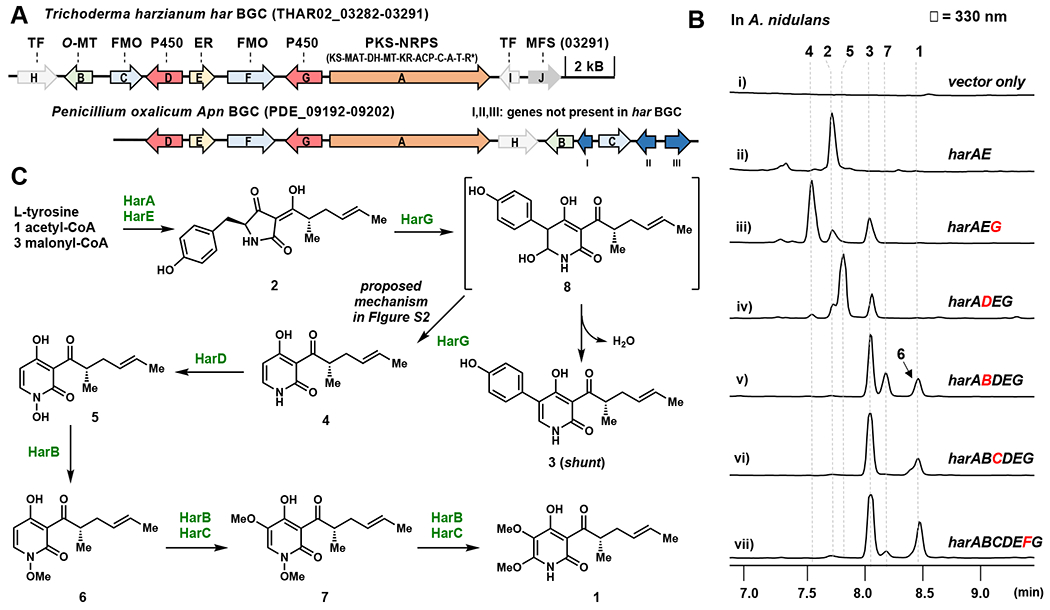
Putative biosynthetic pathway of harzianopyridone 1. (A) The har and apn BGCs. Abbreviations: KS: ketosynthase; MAT: malonyl-CoA:ACP acyltransferase; DH: dehydratase; MT: methyltransferase; KR: ketoreductase; ACP: acyl-carrier protein; C: condensation; A: adenylation; T: thiolation; and R*: Dieckmann cyclization; TF: transcriptional factor; MFS: major facilitator superfamily transporter; (B) HPLC analysis of metabolites produced by the heterologous host A. nidulans. For each trace, gene indicated in red represents addition from the previous construct. Other combinations are in Figure S1. (C) Proposed biosynthetic pathway of 1.
We investigated the metabolites produced by har BGC using Aspergillus nidulans A1145 ΔEM19 as a heterologous expression host (Figure 2B). When HarA-HarF were coexpressed, we detected biosynthesis of several new metabolites (Figure 2B, i and vii). We confirmed that indeed 1 is biosynthesized at 0.5 mg/L (Figures S19–S23, Table S4). Also produced are 5-phenyl-2-pyridone 3 at 5 mg/L (Figures S29–S33, Table S6), and two minor metabolites 6 and 7 that are N1-methoxylated 2-pyridones. Whereas 6 is not substituted at C5 and C6 (2.5 mg/L, Figures S44–S48, Table S9), 7 contains a C5-methoxy group (3 mg/mL, Figures S49–S53, Table S10). The presence of N-methoxy in 6 and 7 was surprising considering 1 does not contain this modification, although this agrees with the role of the predicted N-hydroxylase HarD.
Different combinations of har genes were then expressed in A. nidulans to probe individual enzyme functions (Figures 2B and S1). When HarA and HarE were coexpressed, the tetramic acid 2 was produced at 7.5 mg/L (Figure 2B, ii, Figures S24–S28, Table S5), consistent with the functional assignment of these two enzymes. This shows l-tyrosine is incorporated by the NRPS module of HarA, and a phenol cleavage step20 is required to give 1. When HarG, the proposed P450RE was coexpressed with HarA and HarE, 3 and 4 were formed (Figure 2B, iii). Compound 4 was structurally verified to be the dephenylated 2-pyridone (7.5 mg/L, Figures S34–S38, Table S7). The co-emergence of 3 and 4 suggests that HarG is responsible for both ring expansion and phenyl-cleavage steps. We verified that 3 cannot undergo phenyl cleavage when supplied to A. nidulans expressing HarG, which indicates this compound is a shunt product. Cox and coworkers proposed a radical mechanism for the ring expansion of tetramic acid in tenellin biosynthesis, during which the 6-hydroxy-dihydropyridone such as 8 is a proposed intermediate.20 Subsequent dehydration of 8 can produce 3. We propose that if the P450RE heme-iron can be reduced prior to dehydration of 8, the enzyme can further catalyze oxidation of the phenyl ring of 8, which can lead to loss of quinone and give 4 (mechanism in Figure S2).
We next determined which remaining enzymes install the methoxy groups in 4 to give 1. Only coexpression of the P450 HarD with HarAEG led to transformation of 4 to the N-hydroxy pyridone 5 (0.5 mg/L, Figures 2B, iv and S39–S43, Table S8). Further coexpression of O-MT HarB led to the emergence of 6 and 7 (Figure 2B, v). While methylation of 5 to 6 fits with the predicted function of HarB, methoxylation of C5 in 6 to give 7 is not expected. Based on yeast biotransformation and in vitro assays described below, we conclude formation of 7 here may be due to crosstalk with A. nidulans endogenous enzymes. Finally, when the FMO HarC was expressed together, formation of 1 was observed (Figure 2B, vi). The other FMO HarF is not essential in the reconstitution of 1, which is in agreement with the observed ancillary role of the homologs in other pyridone pathways.20 Therefore, HarABCDEG represents the minimal set of enzymes needed to biosynthesize 1 (Figure 2C). Using the har BGC as a guide, we identified homologous clusters in fungal genomes (Figure S18), including the apn BGC from Penicillium oxalicum, the known atpenin producer (Figure 2A, Table S1).4,21 The apn BGC contains additional genes possibly involved in the chlorination steps. Indeed, heterologous expression of the same set of enzymes (ApnABCDEG) in A. nidulans produced atpenin B at 0.3 mg/L (Figure S17, Table S11).
To examine whether 6 and 7 are pathway intermediates, we performed biotransformations using Saccharomyces cerevisiae (Table 1, Figures S3–S6). Compounds 4–7 were individually fed to yeast expressing combinations of HarB, HarC and HarD. All four compounds are bona fide biosynthetic intermediates, as each can be biotransformed into 1 (entries 3, 7, 11 and 14). Compound 4 was oxidized to 5 by HarD (entry 1), which was methylated by HarB to form 6 (entries 2 and 5). N-methoxylation is required for the formation of 1, as 4 was not consumed by HarB and HarC without HarD (entry 4), nor was 5 oxidized in the presence of HarC only (entry 8). The biotransformation of 5 into 1 by HarB and HarC demonstrates that HarB is responsible for all O-methylation reactions, while HarC is required for both C5 and C6 hydroxylations. The requirement of HarC in transforming 6 to 7 and 1 suggests emergence of 7 in A. nidulans expressing ABDEG (Figure 2B, v) was likely due to endogenous monooxygenases. When 6 was fed to yeast expressing only HarC, we detected a new compound 10 with m/z (+) 286 (entry 9) (Figure S5), which was proposed to be a C5, C6 di-hydroxylated dihydropyridone (Figure 3). We were unable to characterize 10 since it was readily degraded during purification. Lastly when 7 was fed to yeast expressing only HarC, no product could be identified although 7 was consumed (entry 12), indicating formation of an intermediate that degrades in the absence of HarB.
Table 1.
Yeast biotransformation of biosynthetic intermediatesa
| entry | substrate | HarD | HarB | HarC | product(s) |
|---|---|---|---|---|---|
| 1 | 4 | + | − | − | 5 |
| 2 | 4 | + | + | − | 6 |
| 3 | 4 | + | + | + | 1 (>95%), 7 |
| 4 | 4 | − | + | + | No conversion |
| 5 | 5 | − | + | − | 6 |
| 6 | 5 | + | + | − | 6 |
| 7 | 5 | − | + | + | 1 (>95%), 7 |
| 8 | 5 | − | − | + | No conversion |
| 9 | 6 | − | − | + | 10 |
| 10 | 6 | − | + | − | No conversion |
| 11 | 6 | − | + | + | 1 (>90%), 7, 10 |
| 12 | 7 | − | − | + | No conversionb |
| 13 | 7 | − | + | − | No conversion |
| 14 | 7 | − | + | + | 1 |
All substrates fed at 33 μM to S. cerevisiae expressing the indicated combination of enzymes.
the substrate was consumed, but no products were detectable by LCMS.
Figure 3.

Proposed functions of FMO HarC and O-MT HarB in biosynthesis of 1.
To investigate the iterative activities of HarB and HarC, we performed in vitro assays using purified enzymes (Figure S7). Starting with 6, HarB and HarC together were able to produce 1, with a small amount of 7 detected (Figure S8). The reaction requires both NADPH and SAM (Figure S9). The NADPH is required for regeneration of reduced flavin after the hydroxylation reaction. Adding HarC alone to 6 led to decrease in 6 and formation of 10. In vitro conversion of 7 to 1 by HarB and HarC was complete within 20 minutes (~20 turnovers) as shown in Figure S10. 7 was nearly completely consumed by HarC, with no detectable product. The enzyme-catalyzed transformations of 6 and 7 to 1 are oxygen dependent, as reactions performed under anaerobic conditions did not lead to product formation (Figures S12–S13). To investigate the origin of the C5 and C6 oxygen atoms in 1, we performed the enzyme assays in the presence of H218O. We observed the C7 ketone oxygen can exchange with H218O and lead to +2 mu (Figure S14B). Therefore, after each reaction was completed in H218O, the solvent was lyophilized and the samples were re-equilibrated in in H216O before MS analysis (Figure S14A). From 6 to 1, we observed a +2 increase in MW of 1, which indicates one of the C5 and C6 oxygens in 1 is derived from H2O (Figure S15). The +2 increase was also observed in the conversion of 7 to 1 (Figure S16). These labeling results implicate that the first oxygen atom introduced into 6 to form 7 is from molecular oxygen, while the second oxygen introduced to convert 7 to 1 is from water instead of O2 or a methoxy migration.
Our analysis of the biosynthesis of 1 has revealed unexpected biosynthetic logic and raises mechanistic questions (Figure 3). For example, installation of the N-OMe group followed by its removal in the last step is an interesting strategy. One could propose this is a protective group to prevent N-methylation by the iterative HarB. However, assays in which 4 or 1 was incubated with HarB did not result in N-methylation. Alternatively, we propose N-methoxy group serves as a directing group that increases nucleophilic character of the nitrogen through the alpha effect.22 The increased nucleophilicity of N1 promotes electrophilic aromatic substitution (EAS) of 6 at C5 to give 9, using flavin hydroperoxide as an electrophile. In the absence of HarB, 9 can form 10, either via direct water attack at C6 or through an epoxide intermediate 11. HarB could facilitate the aromatization of 9 to 12, followed by C5-O-methylation to give 7.
Following EAS, the N-methoxy directing group can be removed as methanol via a reaction analogous to the Bamberger rearrangement.23 It remains unresolved how the redox neutral conversion from 7 to 1 requires aerobic conditions, while the oxygenation originates from water. As shown in Figure 3, an active site nucleophile (Nu), which we proposed could be flavin peroxide (Fl-OO−) to account for the O2 requirement, can attack C5 to drive the ejection of methanol and yield 13. Elimination of the Nu can form the labile cation 14. HarB may facilitate the attack of water at C5 to form the hemiacetal 15, followed by a semi-pinacol like rearrangement to 16. 16 can rearomatize to 17 and be subjected to C5-O-methylation by HarB to give 1. It may also be possible for a direct water attack at C6 of 14, followed by aromatization and C6-methylation to give 1.
In summary, our work revealed the enzymatic steps in the biosynthesis of 1 and related penta-substituted pyridines. Notably, four of the six enzymes (HarA, G, B and C) perform iterative catalysis, which underscores the highly programmed and unpredictable functions of fungal biosynthetic enzymes.24
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the NIH R01AI141481to YT and KNH. We thank Mengting Liu for help with processing of NMR spectra. We also thank Alec Jaeger and Shih-Wei Chuang for their help in setting up the enzyme assays under anaerobic conditions.
Footnotes
Supporting Information.
This material is available free of charge via the Internet at http://pubs.acs.org. Experimental procedures, chromatograms, and spectroscopic data.
Conflict of Interest: Yi Tang is a co-founder of Hexagon Biosciences, a biotechnology company with interests in natural product discovery.
REFERENCES
- (1).Kluckova K; Bezawork-Geleta A; Rohlena J; Dong L; Neuzil J Mitochondrial Complex II, a Novel Target for Anti-Cancer Agents. Biochim. Biophys. Acta BBA - Bioenerg 2013, 1827, 552–564. [DOI] [PubMed] [Google Scholar]
- (2).Mori M; Nonaka K; Masuma R; Ōmura S; Shiomi K Helminth Electron Transport Inhibitors Produced by Fungi In Physiology and Genetics: Selected Basic and Applied Aspects; Anke T, Schüffler A, Eds.; The Mycota; Springer International Publishing: Cham, 2018; pp 297–329. [Google Scholar]
- (3).Cutler HG; Jacyno JM Biological Activity of (−)-Harzianopyridone Isolated from Trichoderma Harzianum. Agric. Biol. Chem 1991, 55, 2629–2631. [Google Scholar]
- (4).Omura S; Tomoda H; Kimura K; Zhent D-Z; Kumagai H; Igarashi K; Imamura N; Takahashi Y; Tanaka Y; Iwai Y Atpenins, new antifungal antibiotics produced by Penicullium SP. J. Antibiot. (Tokyo) 1988, 41, 1769–1773. [DOI] [PubMed] [Google Scholar]
- (5).Miyadera H; Shiomi K; Ui H; Yamaguchi Y; Masuma R; Tomoda H; Miyoshi H; Osanai A; Kita K; Ōmura S Atpenins, Potent and Specific Inhibitors of Mitochondrial Complex II (Succinate-Ubiquinone Oxidoreductase). Proc. Natl. Acad. Sci 2003, 100, 473–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Horsefield R; Yankovskaya V; Sexton G; Whittingham W; Shiomi K; Ōmura S; Byrne B; Cecchini G; Iwata S Structural and Computational Analysis of the Quinone-Binding Site of Complex II (Succinate-Ubiquinone Oxidoreductase) A mechanism of electron transfer and proton conduction during ubiquionone reduction. J. Biol. Chem 2006, 281, 7309–7316. [DOI] [PubMed] [Google Scholar]
- (7).Jacob Jessen H; Gademann K 4-Hydroxy-2-Pyridone Alkaloids: Structures and Synthetic Approaches. Nat. Prod. Rep 2010, 27, 1168–1185. [DOI] [PubMed] [Google Scholar]
- (8).Trecourt F; Mallet M; Mongin O; Queguiner G Total Synthesis of (.+−.)-Atpenin B. An Original “Clockwise” Functionalization of 2-Chloropyridine. J. Org. Chem 1994, 59, 6173–6178. [Google Scholar]
- (9).Tréecourt F; Mallet M; Mongin O; Quéguiner G First Synthesis of (±)-Harzianopyridone by Metalation of Polysubstituted O-Pyridylcarbamates. J. Heterocycl. Chem 1995, 32 , 1117–1124. [Google Scholar]
- (10).Ohtawa M; Ogihara S; Sugiyama K; Shiomi K; Harigaya Y; Nagamitsu T; Ōmura S Enantioselective Total Synthesis of Atpenin A5. J. Antibiot. (Tokyo) 2009, 62, 289–294. [DOI] [PubMed] [Google Scholar]
- (11).Krautwald S; Nilewski C; Mori M; Shiomi K; Ōmura S; Carreira EM Bioisosteric Exchange of C -Chloro and Methyl Substituents: Synthesis and Initial Biological Studies of Atpenin A5 Analogues. Angew. Chem. Int. Ed 2016, 55, 4049–4053. [DOI] [PubMed] [Google Scholar]
- (12).Selby TP; Hughes KA; Rauh JJ; Hanna WS Synthetic Atpenin Analogs: Potent Mitochondrial Inhibitors of Mammalian and Fungal Succinate-Ubiquinone Oxidoreductase. Bioorg. Med. Chem. Lett 2010, 20, 1665–1668. [DOI] [PubMed] [Google Scholar]
- (13).Wang H; Huwaimel B; Verma K; Miller J; Germain TM; Kinarivala N; Pappas D; Brookes PS; Trippier PC Synthesis and Antineoplastic Evaluation of Mitochondrial Complex II (Succinate Dehydrogenase) Inhibitors Derived from Atpenin A5. ChemMedChem 2017, 12, 1033–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Eley KL; Halo LM; Song Z; Powles H; Cox RJ; Bailey AM; Lazarus CM; Simpson TJ Biosynthesis of the 2-Pyridone Tenellin in the Insect Pathogenic Fungus Beauveria Bassiana. ChemBioChem 2007, 8, 289–297. [DOI] [PubMed] [Google Scholar]
- (15).Dickinson JM; Hanson JR; Hitchcock PB; Claydon N Structure and Biosynthesis of Harzianopyridone, an Antifungal Metabolite of Trichoderma Harzianum. J. Chem. Soc. Perkin 1 1989, 0, 1885–1887. [Google Scholar]
- (16).Halo LM; Heneghan MN; Yakasai AA; Song Z; Williams K; Bailey AM; Cox RJ; Lazarus CM; Simpson TJ Late Stage Oxidations during the Biosynthesis of the 2-Pyridone Tenellin in the Entomopathogenic Fungus Beauveria Bassiana. J. Am. Chem. Soc 2008, 130, 17988–17996. [DOI] [PubMed] [Google Scholar]
- (17).Ohashi M; Liu F; Hai Y; Chen M; Tang M; Yang Z; Sato M; Watanabe K; Houk KN; Tang Y SAM-Dependent Enzyme-Catalysed Pericyclic Reactions in Natural Product Biosynthesis. Nature 2017, 549, 502–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Bergmann S; Schümann J; Scherlach K; Lange C; Brakhage AA; Hertweck C Genomics-Driven Discovery of PKS-NRPS Hybrid Metabolites from Aspergillus Nidulans. Nat. Chem. Biol 2007, 3, 213–217.. [DOI] [PubMed] [Google Scholar]
- (19).Liu N; Hung Y-S; Gao S-S; Hang L; Zou Y; Chooi Y-H; Tang Y Identification and heterologous production of a benzoyl- primed tricarboxylic acid polyketide intermediate from the zaragozic acid a biosynthetic pathway. Org. Lett 2017. 19, 3560–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Wasil Z; K. Pahirulzaman KA; Butts C; J. Simpson T; M. Lazarus C; J. Cox R One Pathway, Many Compounds: Heterologous Expression of a Fungal Biosynthetic Pathway Reveals Its Intrinsic Potential for Diversity. Chem. Sci 2013, 4, 3845–3856. [Google Scholar]
- (21).Penicillium oxalicum as a producer of atpenin A5 can be found in information provided by Prof. Satoshi Ōmura’s website: http://www.satoshi-omura.info/molecules/atpenina4.html (accessed Mar 17, 2020).
- (22).Jencks WP Catalysis in Chemistry and Enzymology; McGraw-Hill Inc: New York, 1969. [Google Scholar]
- (23).Bamberger E Ueber die Reduction der Nitroverbindungen. Chemische Berichte. 1894, 27, 1347–1350. [Google Scholar]
- (24).Hang L; and Liu N; Tang Y “Coordinated and Iterative Enzyme Catalysis in Fungal Polyketide Biosynthesis.” ACS Catalysis 2016, 6, 5935–5945 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.