Figure 3.
Interpatient Variation in iNSC Gene-Expression Profile Following Reprogramming Protocol
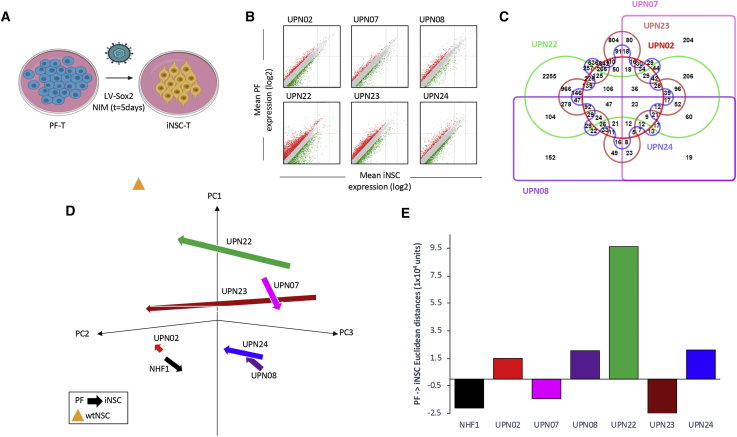
(A) Schematic depiction of therapeutic iNSC transdifferentiation from PF-T. Differential expression analysis (B) showed that our rapid transdifferentiation protocol induced significant alterations in the transcriptomic profile compared to PFs (p < 0.05). A list of genes that changed by 2-fold or greater was compiled for each patient, and an Edward’s Venn diagram was generated from all six lists (C). (D) PCA of all PF and iNSC generated, as well as wild-type NSCs and NHF1-derived fibroblasts and iNSCs, again demonstrated patient-specific variation in reprogramming. The beginning of arrows indicates PF coordinates, and arrowheads indicate iNSC coordinates following reprogramming. (E) Summary graph of mean Euclidean distances between PFs and iNSCs generated from the same unique patient. Vector direction was determined based on principal component 1 (PC1).