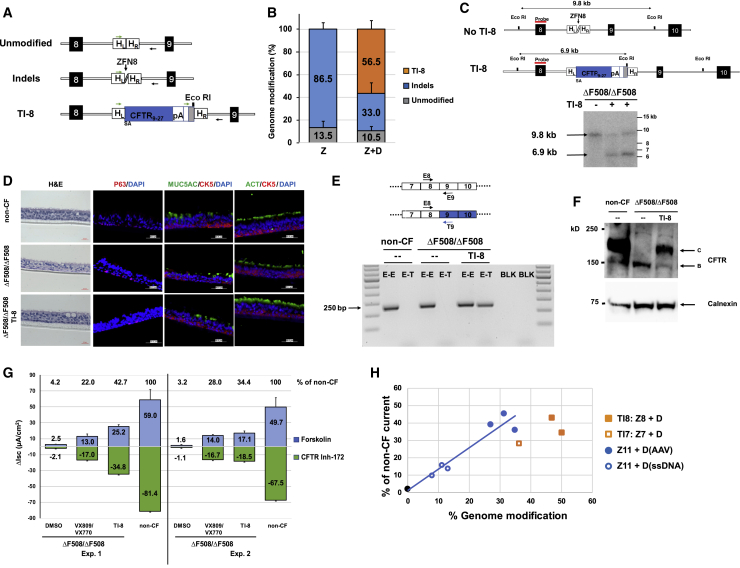
Figure 3.
Efficient SA-CFTR9-27-pA TI into CFTR Intron 8 of ΔF508/ΔF508 Airway Basal Cells
(A) Schematic of site-specific targeted editing of CFTR intron 8. Unmodified and indel diagrams highlight CFTR genomic sequences between exons 8 and 9 (black boxes; not to scale). TI-8 diagram shows intron 8 TI of human codon optimized CFTR9-27 cDNA preceded by a splice acceptor, followed by bovine growth hormone (bGH) pA sequence, and flanked by 313 bp homology left (HL) and 351 bp homology right (HR) intron 8 sequences. Arrows indicate oligos amplifying unmodified, indel, or TI-8 events and used to quantify frequency of each by NGS. (B) The percentage of genome modification (mean ± SD) determined by NGS (Table S3A) from five independent TI-8 experiments (utilizing the AAV-6 donor at either 2 × 106 or 6 × 106 viral genomes [vg]/cell). There was no enhancement of TI efficiency as one increased the amount of donor. Efficiency was measured 4 days after the delivery of ZFNs targeting intron 8 (ZFN8) followed immediately by AAV-6 CFTR9-27 cDNA donor. Z, ZFN8 alone; Z+D, ZFN8 and AAV-6 donor. (C) Southern blot analysis of TI-8. The schematic shows the expected genomic organization of non-targeted (no TI-8) and targeted (TI-8) alleles with the expected sizes resulting from EcoRI digestion. The non-targeted allele yields a 9.8 kb band while the TI-8 allele yields a 6.9 kb band due to the new EcoRI site in the CFTR9-27 cDNA donor construct. (D) H&E staining and immunostaining of well-differentiated airway epithelium in ALI culture. Representative images of cross sections from ΔF508/ΔF508 or TI-8 ΔF508/ΔF508. Immunostaining shows major epithelial cell types: basal cell (CK5 and P63), secretory cells (MUC5AC), and ciliated cells (ACT). 40× images, scale bar, 50 μm. (E) Detection of transgene CFTR9-27 mRNA. Schematic of endogenous and transgene CFTR mRNA is shown here. Primer T9 recognizes codon-optimized transgene exon 9 sequence only (blue) while Primer E9 recognizes endogenous exon 9. A 250 bp E8-T9 RT-PCR amplicon, evidence of the chimeric endogenous-transgene CFTR mRNA, was present only in the TI-8 ΔF508/ΔF508 sample. (F) Restoration of fully glycosylated CFTR protein via TI-8. Western blots of protein lysates harvested at 4 weeks of ALI culture. Band C, representing the mature, fully glycosylated form of CFTR, is present in non-CF, absent in ΔF508/ΔF508 cells, and restored in TI-8 ΔF508/ΔF508. Calnexin, loading control. (G) CFTR functional assay in ALI cultures at 4 weeks. TI-8 ΔF508/ΔF508 cells show the restoration of CFTR function measured as Δlsc (μA/cm2) ± SD and compared with ΔF508/ΔF508 treated with VX-809/VX-770. (H) Restored CFTR activity as function of gene-editing frequency. The blue and black symbols and blue linear regression line are from individual ΔF508/ΔF508 cell experiments with sequence-specific correction of ΔF508 (from Figure 2G). Shown in filled square and open square orange symbols are the results of ΔF508/ΔF508 cell TI-8 and ΔF508/ΔF508 cell TI-7 experiments, respectively.