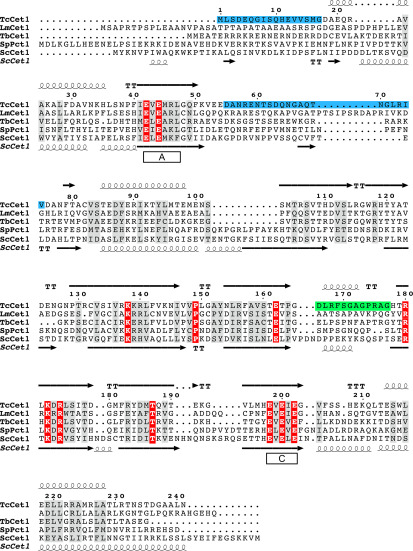
Figure 1.
Structure-based alignment of T. cruzi RNA triphosphatase. The secondary structure of T. cruzi RNA triphosphatase (TcCet1) is shown above the amino acid sequence. TcCet1 amino acids shaded in blue correspond to the segments that are removed in TcCet1(18–243 Δ55–75) protein. The TcCet1 sequence shaded in green (amino acid residues 166–177) corresponds to the segment that interacts with C13H13NO2 and C10H14N4O2 but was disordered in TcCet1(18–243 Δ55–75) manganese and Mn·PPPi–bound structures. The amino acid sequences of TcCet1 is aligned with Leishmania major (LmCet1), T. brucei (TbCet1), Schizosaccharomyces pombe (SpPct1), and S. cerevisiae (ScCet1) RNA triphosphatases. The secondary structure of ScCet1 is indicated below the aligned sequences. Identical side chains found in all polypeptides are highlighted in red. Amino acids with similar side chains are highlighted in gray. The positions of conserved motifs A and C, located within the catalytic domain of the metal-dependent RNA triphosphatases, are indicated. The alignment was prepared by ESPript. The secondary structure assignment was based on the DSSP program.