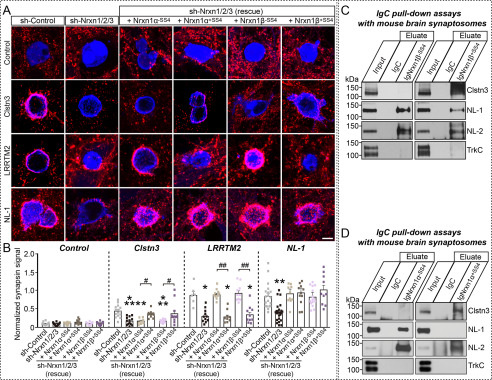
Figure 5.
Selected Nrxn variants are required for Clstn3-mediated induction of presynaptic differentiation. A and B, effects of Nrxn-TKD (sh-Nrnx1/2/3) on the synaptogenic activities of Clstn3, LRRTM2, and NL-1. HEK293T cells expressing the indicated proteins were co-cultured with neurons infected with control lentiviruses (sh-Control) or lentiviruses expressing sh-Nrxn1/2/3, without or with coexpression of the indicated Nrxn1α or Nrxn1β splice variants. Representative images (A) of co-cultures immunostained with antibodies to mVenus or HA (blue) and synapsin I (red). Scale bar: 10 μm (applies to all images). Quantitation (B) of heterologous synapse-formation assays, determined by calculating the ratio of synapsin to EGFP/HA fluorescence signals. Dashed lines correspond to control values used as a baseline. Data are mean ± S.E. (***, p < 0.001; **, p < 0.01; *, p < 0.05; ##, p < 0.01; and #, p < 0.05; nonparametric Kruskal-Wallis test with Dunn's post hoc test; sh-Control/Control, n = 10; sh-Nrxn1/2/3/Control, n = 10; sh-Nrxn1/2/3 (rescue + Nrxn1α+SS4)/Control, n = 10; sh-Nrxn1/2/3 (rescue + Nrxn1α-SS4)/Control, n = 9; sh-Nrxn1/2/3 (rescue + Nrxn1β+SS4)/Control, n = 11; sh-Nrxn1/2/3 (rescue + Nrxn1β-SS4)/Control, n = 10; sh-Control/Clstn3, n = 15; sh-Nrxn1/2/3/Clstn3, n = 13; sh-Nrxn1/2/3 (rescue + Nrxn1α+SS4)/Clstn3, n = 8; sh-Nrxn1/2/3 (rescue + Nrxn1α-SS4)/Clstn3, n = 11; sh-Nrxn1/2/3 (rescue + Nrxn1β+SS4)/Clstn3, n = 11; sh-Nrxn1/2/3 (rescue + Nrxn1β-SS4)/Clstn3, n = 13; sh-Control/LRRTM2, n = 6; sh-Nrxn1/2/3/LRRTM2, n = 11; sh-Nrxn1/2/3 (rescue + Nrxn1α+SS4)/LRRTM2, n = 10; sh-Nrxn1/2/3 (rescue + Nrxn1α-SS4)/LRRTM2, n = 9; sh-Nrxn1/2/3 (rescue + Nrxn1β+SS4)/LRRTM2,n = 10; sh-Nrxn1/2/3 (rescue + Nrxn1β-SS4)/LRRTM2, n = 9; sh-Control/NL-1, n = 19; sh-Nrxn1/2/3/NL-1, n = 25; sh-Nrxn1/2/3 (rescue + Nrxn1α+SS4)/NL-1, n = 10; sh-Nrxn1/2/3 (rescue + Nrxn1α-SS4)/NL-1, n = 12; sh-Nrxn1/2/3 (rescue + Nrxn1β+SS4)/NL-1, n = 15; and sh-Nrxn1/2/3 (rescue + Nrxn1β-SS4)/NL-1, n = 10). p values for Control conditions: sh-Control versus sh-Nrxn1/2/3, p > 0.9999; sh-Nrxn1/2/3 versus sh-Nrxn1/2/3 (rescue + Nrxn1α-SS4), p > 0.9999; sh-Nrxn1/2/3 versus sh-Nrxn1/2/3 (rescue + Nrxn1α+SS4), p > 0.9999; sh-Nrxn1/2/3 versus sh-Nrxn1/2/3 (rescue + Nrxn1β-SS4), p > 0.9999; sh-Nrxn1/2/3 versus sh-Nrxn1/2/3 (rescue + Nrxn1β+SS4), p > 0.9999. p values for Clstn3 condition: sh-Control versus sh-Nrxn1/2/3, p = 0.003; sh-Control versus sh-Nrxn1/2/3 (rescue + Nrxn1α-SS4), p = 0.0026; sh-Control versus sh-Nrxn1/2/3 (rescue + Nrxn1α+SS4), p > 0.9999; sh-Control versus sh-Nrxn1/2/3 (rescue + Nrxn1β-SS4), p = 0.0003; sh-Control versus sh-Nrxn1/2/3 (rescue + Nrxn1β+SS4), p > 0.9999; sh-Nrxn1/2/3 (rescue + Nrxn1α-SS4) versus sh-Nrxn1/2/3 (rescue + Nrxn1α+SS4), p = 0.0427; sh-Nrxn1/2/3 (rescue + Nrxn1β-SS4) versus sh-Nrxn1/2/3 (rescue + Nrxn1β+SS4), p = 0.0123. p values for LRRTM2 condition: sh-Control versus sh-Nrxn1/2/3, p = 0.0252; sh-Control versus sh-Nrxn1/2/3 (rescue + Nrxn1α-SS4), p > 0.9999; sh-Control versus sh-Nrxn1/2/3 (rescue + Nrxn1α+SS4), p = 0.325; sh-Control versus sh-Nrxn1/2/3 (rescue + Nrxn1β-SS4), p > 0.9999; sh-Control versus sh-Nrxn1/2/3 (rescue + Nrxn1β+SS4), p = 0.0285; sh-Nrxn1/2/3 (rescue + Nrxn1α-SS4) versus sh-Nrxn1/2/3 (rescue + Nrxn1α+SS4), p = 0.007; sh-Nrxn1/2/3 (rescue + Nrxn1β-SS4) versus sh-Nrxn1/2/3 (rescue + Nrxn1β+SS4), p = 0.0021. p values for NL-1 condition: sh-Control versus sh-Nrxn1/2/3, p = 0.0015; sh-Control versus sh-Nrxn1/2/3 (rescue + Nrxn1α-SS4), p > 0.9999; sh-Control versus sh-Nrxn1/2/3 (rescue + Nrxn1α+SS4), p > 0.9999; sh-Control versus sh-Nrxn1/2/3 (rescue + Nrxn1β-SS4), p > 0.9999; sh-Control versus sh-Nrxn1/2/3 (rescue + Nrxn1β+SS4), p > 0.9999; sh-Nrxn1/2/3 (rescue + Nrxn1α-SS4) versus sh-Nrxn1/2/3 (rescue + Nrxn1α+SS4), p > 0.9999; sh-Nrxn1/2/3 (rescue + Nrxn1β-SS4) versus sh-Nrxn1/2/3 (rescue + Nrxn1β+SS4), p > 0.9999. C and D, pulldown assays in solubilized mouse synaptosomal fractions. Assays were performed using recombinant IgNrxn1β (C), IgNrxn1α (D), or IgC proteins. Equal amounts of bound proteins were analyzed using the antibodies indicated to the right of panels. Input, 5%.