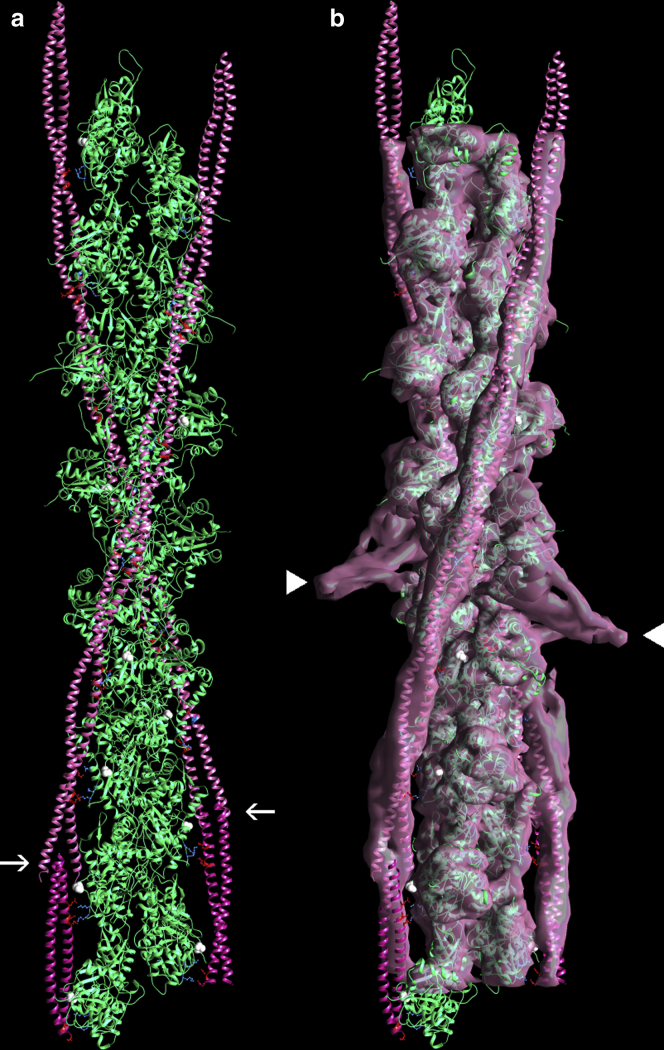
Figure 3.
Model of actin-tropomyosin following molecular dynamics. (a) The actin-tropomyosin structure shown was started from a canonical model of tropomyosin fitted to the cryo-EM three-dimensional reconstruction (EMDB EMD-0729) and then threaded into the structure of the tropomyosin overlapping domain and troponin-T complex common to (7,24). MD was run for 30 ns. The tropomyosin coiled coil remained intact throughout simulation, and side-chain interactions noted in Table 1 also were stable during MD. Representative trajectory (a) fitted within EMD-0729 is shown (b), with the same color coding and highlighting as in Figs. 1 and 2. The trajectory snapshot was taken from the 27 to 30 ns time period of simulation and is a frame that had the lowest root mean-square deviation from the average structure over that same time period (calculated by backbone atom superimposition). PDB coordinates are provided as Data S1. To see this figure in color, go online.