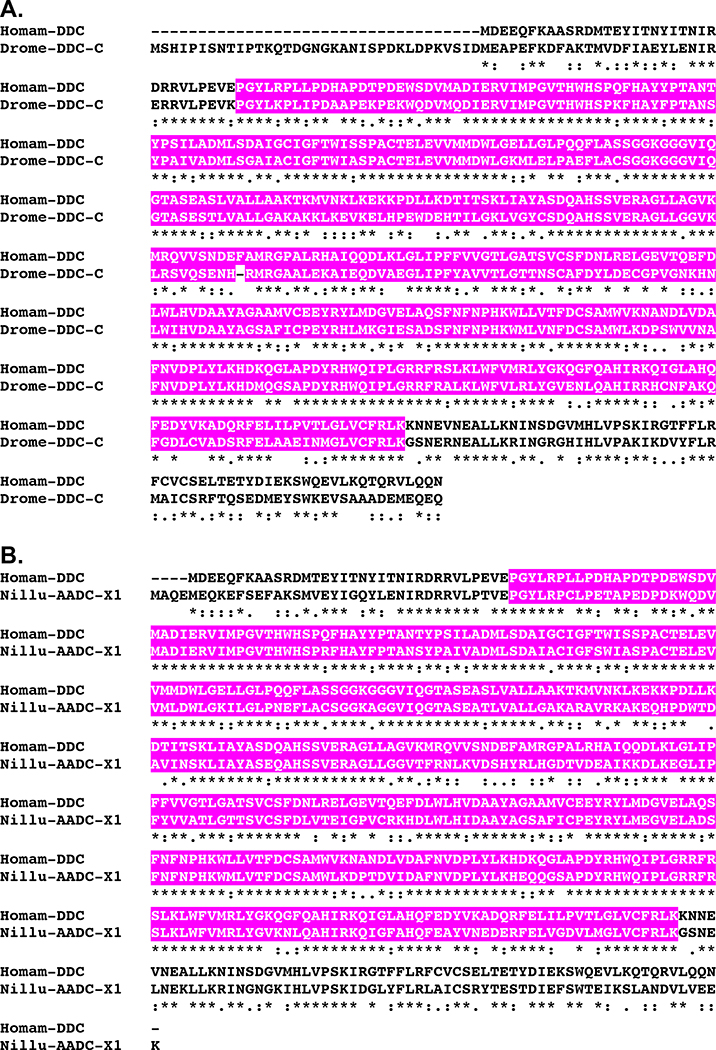
Figure 6.
Alignment of Homarus americanus and related DOPA decarboxylases. (A) MAFFT alignment of H. americanus DOPA decarboxylase (Homam-DDC) and Drosophila melanogaster DOPA decarboxylase isoform C (Drome-DDC-C; Accession No. AAF53762; Adams et al., 2000). (B) MAFFT alignment of Homam-DDC and Nilaparvata lugens aromatic-L-amino-acid decarboxylase, a synonym for DDC, isoform X1 (Nillu-AADC-X1; Accession No. XP_022191934; unpublished direct GenBank submission). In the line immediately below each sequence grouping, “*” indicates identical amino acid residues, while “:” and “.” denote amino acids that are similar in structure between sequences. In this figure, pyridoxal-dependent decarboxylase conserved domains identified by Pfam analyses are highlighted in pink.