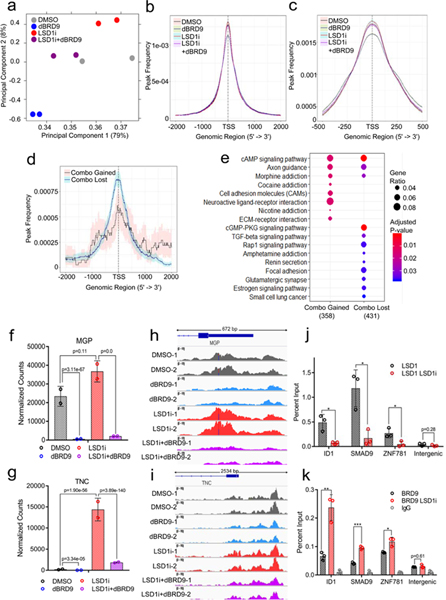
Fig. 8. BRD9 is required to de-repress the expression of LSD1 target genes.
a-i. Inhibition of LSD1 and BRD9 alters chromatin accessibility of an overlapping set of genes in an antagonistic manner (Source Data 8). Two biological replicates of ATAC-seq with MKL-1 cells treated with DMSO (Control), GSK-LSD1 (LSD1i, 0.1 μM), dBRD9 (0.1 μM) or both LSD1i and dBRD9 (LSD1i+dBRD9) for three days were performed. a. The PCA plot displays global changes in chromatin accessibility following the mentioned treatments. b. Peak count frequencies in the transcription start sites (TSSs, +/− 2,000 bp) for the mentioned conditions are shown. The shaded areas display 95% confidence intervals. c. Peak count frequencies of ATAC-seq with the mentioned conditions are shown (TSSs +/− 500). d. Frequencies of the differentially enriched peaks between LSD1i and LSD1i+dBRD9 are shown. ‘Combo_gained’ displays frequencies of peaks gained in the LSD1i+dBRD9 combination compared to LSD1i, whereas ‘Combo_lost’ shows frequencies of peaks lost in the combination. e. GOTERM biological process pathways enriched with the genes annotated with Combo_gained (n=358) and Combo_lost (n=431) peaks are shown. The enrichment test was performed on hypergeometric distribution using the clusterProfiler R package49 and the p-values were adjusted by FDR49. f-g. DESeq2 normalized counts of MGP (f) and TNC (g) from the RNA-seq of MKL-1 cells treated with DMSO (DMSO), GSK-LSD1 (LSD1i, 0.1 μM), dBRD9 (0.1 μM) or both LSD1i and dBRD9 (LSD1i+dBRD9) are shown (Fig. 7c-e). Differentially expressed genes were found by comparing each condition with DESeq242, and p-values were adjusted by Benjamini-Hochberg. h-i. ATAC-seq peaks in the promoters of MGP (h) and TNC (i) in the mentioned conditions with two biological replicates (_1 and _2) are shown. j-k. ChIP-qPCR reveals that LSD1 binding decreases (j), whereas BRD9 binding increases during LSD1 inhibition (k). MKL-1 cells were treated with GSK-LSD1 for three days. Data are shown as mean ± SD; * P<0.05, ** P<0.005, and ***P<0.0005. Data are shown as mean of n=3 ± SD; two-sided t-test, *P<0.05; **<0.005; ***<0.0005.