Figure 2.
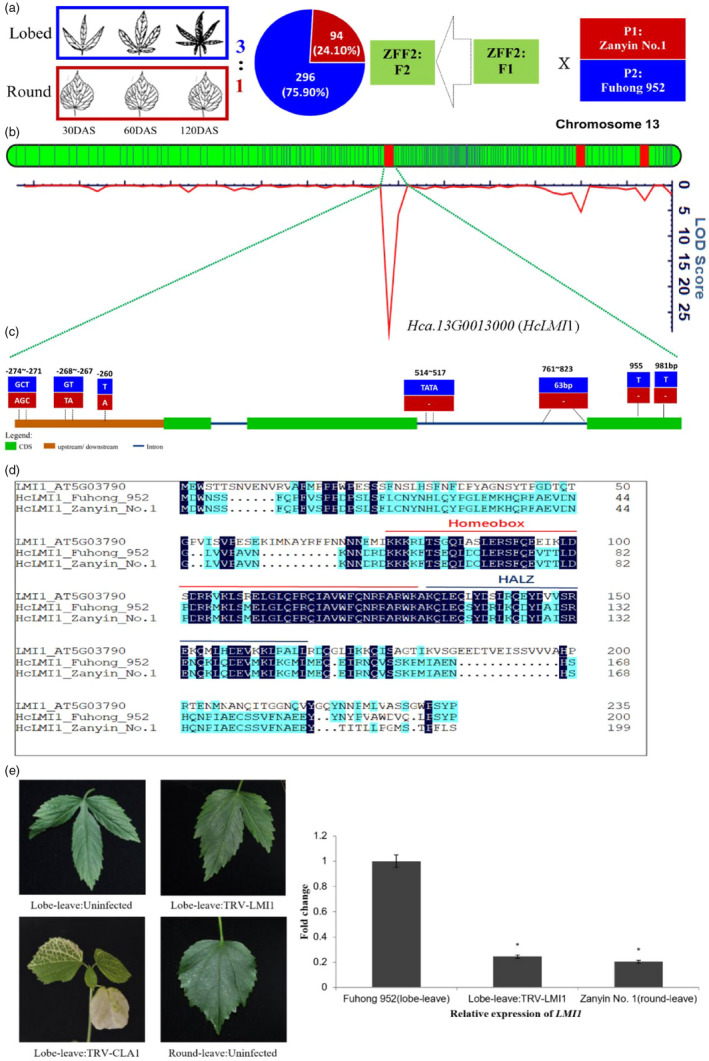
Molecular mapping of leaf shape gene (HcLMI1). (a) The 3:1 segregation ratio of lobed‐leaf: round‐leaf in the F2 population indicated a major gene controlling this trait with lobed‐leaf as dominate and round‐leaf recessive (Zhang et al. 2019). (b) LOD score for leaf shape gene mapping, based on a high‐density SNP genetic map, shown in Table S9. (c) Gene structure and variation in a candidate leaf shape gene HcLMI1. Exons and introns are represented by boxes and lines, respectively. The position of the causal micro‐structure variation is marked. Red and blue boxes represent P1‐’Zanyin No. 1’ and P2‐‘Fuhong 952’, respectively. (d). Amino acid comparison of Hc LMI1 between P1‐’Zanyin No. 1’ and P2‐‘Fuhong 952’. The red line: Homeobox domain. The blue line: HALZ domain. e. Representative leaves from VIGS experiment showing the reversion to round‐like leaf shape (Lobed‐leaf: TRV‐LMI1) in the HcLMI1 silencing treatment; Albino phenotype of HcCLA1 silenced kenaf plants represents successfully establishment of tobacco rattle virus‐induced genes silencing (TRV‐VIGS); Relative transcript levels of candidate genes in the HcLMI1 silenced and control Lobe‐leave plants (n = 3) confirmed the effective knockdown of LMI1. Asterisks represent statistically significant differences as determined by unpaired t tests at P < 0.05.
