Figure 6.
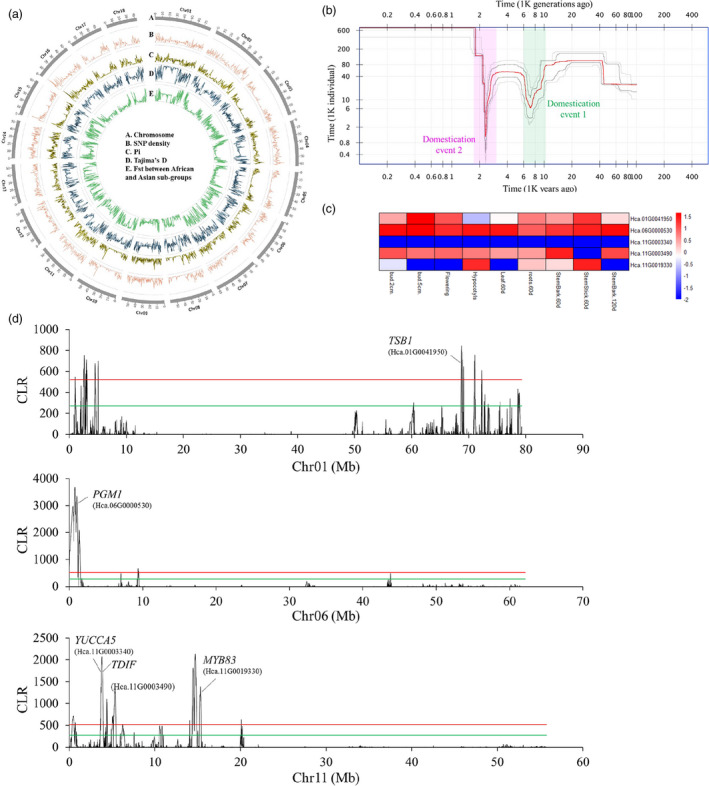
Genomic‐wide genetic diversity and selection signals under fibre improvement of H. cannabinus. a. Genome wide genetic diversity among core kenaf cultivars. From the outer to the centre present (A) chromosome position; (B) SNP density; (C) nucleotide diversity Pi; (D) Tajima’s D; (E) F st between African and Asian subgroups. b. Historical effective population size (Ne) for domesticated kenaf population beginning from 101 Kya years ago to present. The plot showing kenaf population has undergone domesticated bottlenecks during two periods, including one ancient domestication event at ~10 k (green shade) years ago and a recent domestication event at ~2.3 k (pink shade) years ago. The estimate is median (red line) from 200 bootstrap replicates with 87.5% and 97.5% confidence intervals (four grey lines). (c) The fibre synthesis and metabolism‐related genes under selective sweeps regions were marked in the graphs. The fibre related genes MYB83, TDIF, YUCCA5, PGM1 and TSB1 may be under artificial selection during human domestication of kenaf fibres. The red solid line indicates the candidate regions above the 1% (the green solid line indicates 5%) cut‐off outlier with significant deviations from neutrality. d. Expression profile of five fibre related genes (MYB83, TDIF, YUCCA5, PGM1 and TSB1) under artificial selection during human domestication of bast fibres.
