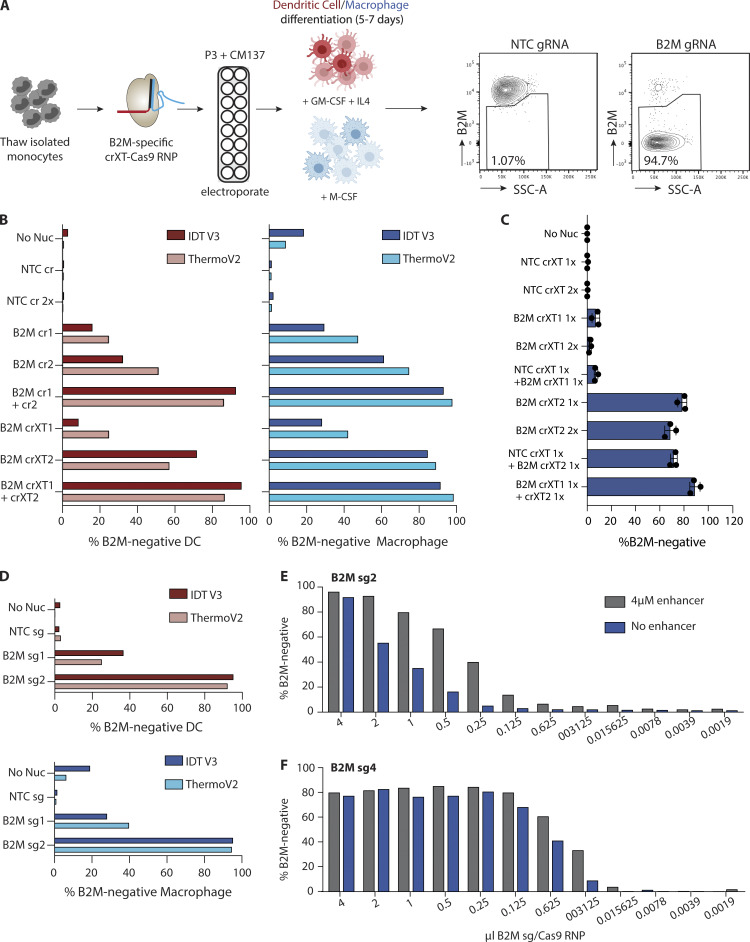
Figure 2.
Population-level gene editing in human monocyte-derived DCs and macrophages. (A) Workflow for B2M-specific KO in human monocyte-derived dendritic and macrophage cultures. Representative flow cytometry plots show gating strategy for using Cas9-RNPs loaded with NTC gRNA to determine B2M-negative cells in each cell population. (B) B2M KO efficiency in monocyte-derived DC (pink, left bar graph) and macrophage (right, blue bars) cells nucleofected with distinct B2M targeting sequences in either crRNA (B2M cr1, B2M cr2) or crRNAXT (B2M crXT1, B2M crXT2) format, or NTC complexed with IDT V3 Cas9 (dark bars) or Thermo Fisher TruCut V2 Cas9 (light bars). Data are from one experiment. (C) B2M KO efficiency in monocyte-derived macrophages nucleofected with IDT V3 Cas9-RNPs loaded with two different crRNAXTs (crXT1 and crXT2) targeting B2M or NTC (NTC crXT). Cas9-RNPs were added individually or in combination. Each Cas9-RNP is labeled with 1× or 2× to indicate the relative molar quantity nucleofected into the cells. Data are presented as mean ± SD (n = 3). (D) Same as B, but gRNA-Cas9-RNPs were loaded with sgRNAs (B2M sg1 and sg2) instead of crRNAs or crRNAXTs. (E and F) Dose–response curve of B2M KO efficiency in monocyte-derived macrophages. IDT V3 Cas9-RNPs loaded with two different sgRNAs targeting B2M (sg2 or sg4) were nucleofected into cells at the indicated quantities. Cas9-RNPs were complexed and delivered with and without 4 µM of IDT electroporation enhancer. Data in B–D are representative of two independent experiments (n = 1 donor per experiment). Data in E and F are from one experiment (n = 1 donor). (B–E) No nucleofection (No Nuc) cells or cells nucleofected with NTC crRNA, crRNAXT, or sgRNA-Cas9-RNPs were used as controls. Buffer P3, CM-137 condition was used for all Cas9-RNP delivery.