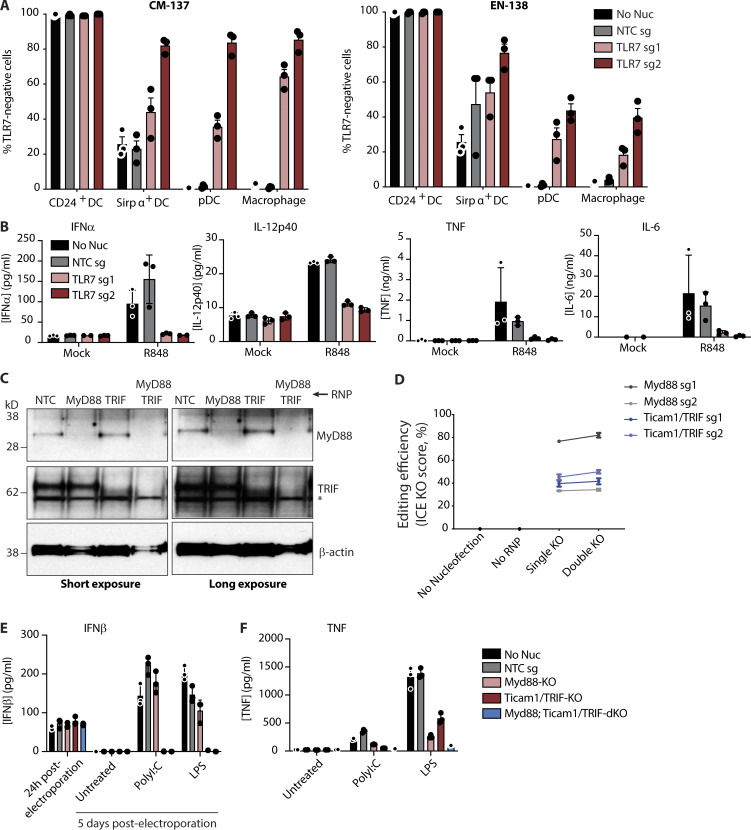
Figure 3.
Disruption of single or multiple genes in murine BMDCs and BMDMs to study TLR signaling. (A) Percentage of cells that were TLR7 negative from BM-derived CD24+, Sirpα+ DCs, pDCs, and macrophages nucleofected with Cas9-RNPs loaded with an NTC or two different Tlr7-specific sgRNAs (sg1 and sg2). Left: Buffer P3, Program CM-137; Right: Buffer P3, Program EN-138. TLR7-negative cells were assayed by intracellular flow cytometry. Data are presented as mean ± SEM (n = 3, biological triplicates) and representative of two independent experiments. (B) Cytokine levels measured by Luminex in supernatant from BMDC culture (combined cell types) in A after stimulation with mock or 800 ng/ml of the TLR7 agonist R848 for 17 h. Data are presented as mean ± SD (n = 3, technical triplicates) and are representative of two independent experiments. (C) Representative Western blots depicting MyD88 or TRIF knockdown by sgRNA-Cas9-RNP in murine BMDMs. (D) Assessment of gene editing efficiency by Sanger sequencing 5 d after electroporation. Data are presented as mean ± SEM (n = 3) and representative of one experiment. (E) ELISA measurement of IFNβ levels in cell culture medium of BMDMs 24 h after electroporation and 5 d after electroporation treated as described. (F) ELISA measurements of TNF in cell culture supernatant following stimulation with the indicated ligand for 18 h. Data in E and F are presented as mean ± SD (n = 3) and representative of three independent experiments.