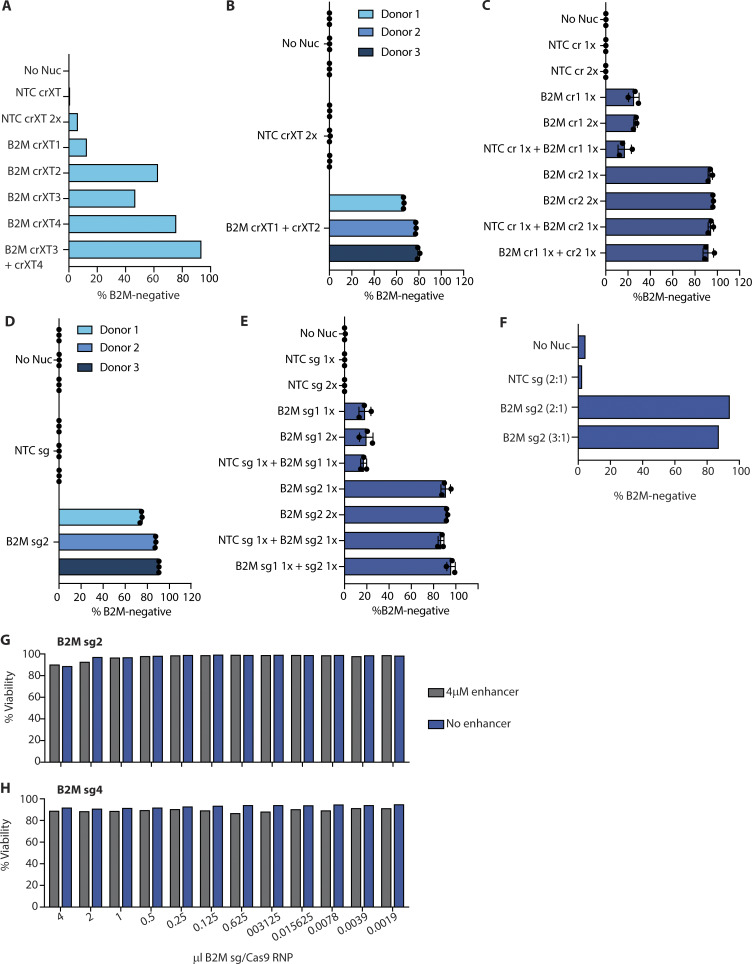
Figure S3.
Supporting data for population-level gene disruption in human monocyte-derived DCs and macrophages. (A) B2M KO efficiency in monocyte-derived macrophages nucleofected with four different crRNAXTs (crXT1, crXT2, crXT3, and crXT4) targeted against B2M and complexed with Cas9. Data are from one experiment. (B) B2M KO efficiency in monocyte-derived macrophages nucleofected with two unique crRNAXTs (crXT1 and crXT2) targeted against B2M and complexed with Cas9. Data from three different donors are displayed, each performed in triplicate (n = 3). (C) B2M KO efficiency in monocyte-derived macrophages nucleofected with indicated crRNAs targeted against B2M or NTC and complexed with Cas9. (D) B2M KO efficiency in monocyte-derived macrophages nucleofected with a single sgRNA (sg2) and complexed with Cas9. Data from three different donors are displayed, each performed in triplicate (n = 3), and are displayed as mean ± SD. (E) B2M KO efficiency in monocyte-derived macrophages nucleofected with IDT V3 containing Cas9-RNPs loaded with two different sgRNAs (sg1 and sg2) targeting B2M or a NTC sgRNA (NTC sg). Cas9-RNPs were added individually or in combination. Each Cas9-RNP is labeled with 1× or 2× to indicate the relative molar quantity nucleofected into the cells. Data in C and E are mean ± SD (n = 3) and representative of two independent experiments. (F) B2M KO efficiency in monocyte-derived macrophages for IDT V3 Cas9-RNPs loaded with NTC sgRNA or B2M sgRNA2 and complexed with an sgRNA:Cas9 molar ratio of 2:1 or 3:1. (G and H) Cell viability in monocyte-derived macrophages from samples in Fig. 2, E and F. Data in F–H are from single donors (n = 1) and one individual experiment.