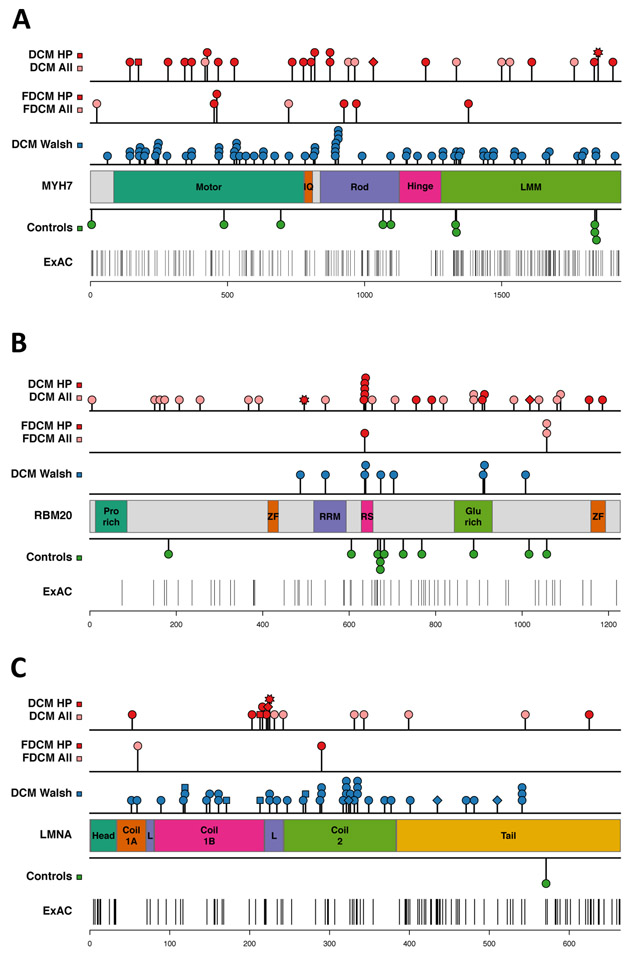
Figure 3.
Rare variant distribution in DCM cases and control subjects. The location of variants in different protein domains is shown for the group A genes: (A) MYH7, (B) RBM20, (C) LMNA. Variants identified in DCM patients in the discovery cohort (top row) and in the familial DCM (FDCM) replication cohort (second row) are shown: all variants (pink, “DCM all”), damaging variants (red, “DCM HP”). Variant types are indicated by: circle =missense, square = splice site change, diamond = frameshift, star = stop codon. The third row shows DCM variants identified by two clinical diagnostic laboratories (“DCM Walsh”)12. Missense variants identified in control subjects are shown below the protein schematic: control subjects in this study (Controls), Exome Aggregation Consortium (ExAC) database. See Supplementary Table S9 for protein domain coordinates and statistical analysis.