Fig. 5. TRIP13 antagonizes REV7 function in the translesion synthesis/FA pathway.
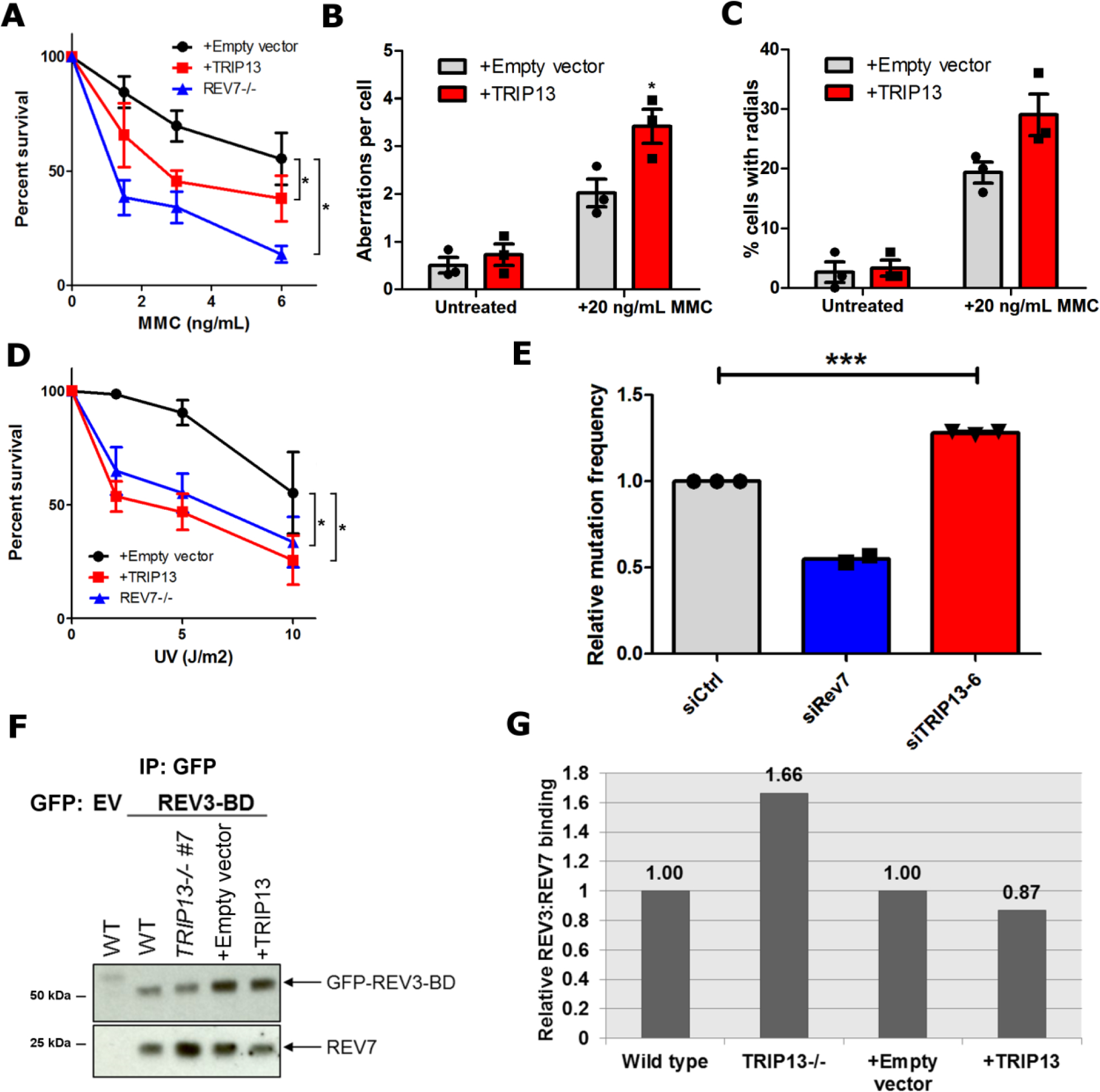
a. 14-day clonogenic survival assay of U2OS wild-type expressing pBabe-empty vector or pBabe-TRIP13 and REV7−/− cell lines treated with the indicated mitomycin C (MMC) doses. n=4 biologically independent experiments (Except for REV7−/−: n=3), Wild-type + Empty vector vs. Wild-type + TRIP13: p = 0.02, Wild-type + Empty vector vs. REV7−/−: p < 0.0001 (2-Way ANOVA). b. Average number of chromosomal aberrations per cell in baseline condition or following treatment with 20 ng/mL of MMC, with or without TRIP13 overexpression. n=3 biologically independent experiments. c. Percentage of cells with radial chromosome formation in baseline condition or following treatment with 20 ng/mL of MMC, with or without TRIP13 overexpression. n=3 biologically independent experiments. d. 14-day clonogenic survival assay of U2OS wild-type expressing pBabe-empty vector or pBabe-TRIP13 and REV7−/− cell lines treated with the indicated UV doses. n=4 biologically independent experiments, Wild-type + Empty vector vs. Wild-type + TRIP13: p < 0.0001, Wild-type + Empty vector vs. REV7−/− : p = 0.002 (2-Way ANOVA). e. Relative UV-induced mutation frequencies in HEK293T cells transfected with nontargeting, REV7- or TRIP13-targeting siRNAs as measured by the SupF assay. n=3 biologically independent experiments (Except for REV7−/−: n=2), siCtrl vs siTRIP13–6: p = 0.0006 (Student’s paired t-test, two-tailed). f. Western blot of GFP IP from U2OS wild type, TRIP13−/−, pBabe and pBabe-TRIP13 cells transfected with GFP-tagged REV3 binding domain (REV3-BD). g. Quantification of western blot in (f). All error bars represent SEM. All immunoblots are representative of at least 2 independent experiments. Statistical source data are provided in Source Data Fig. 5. Unprocessed blots are provided in Unprocessed blots Fig. 5.
