Figure 1.
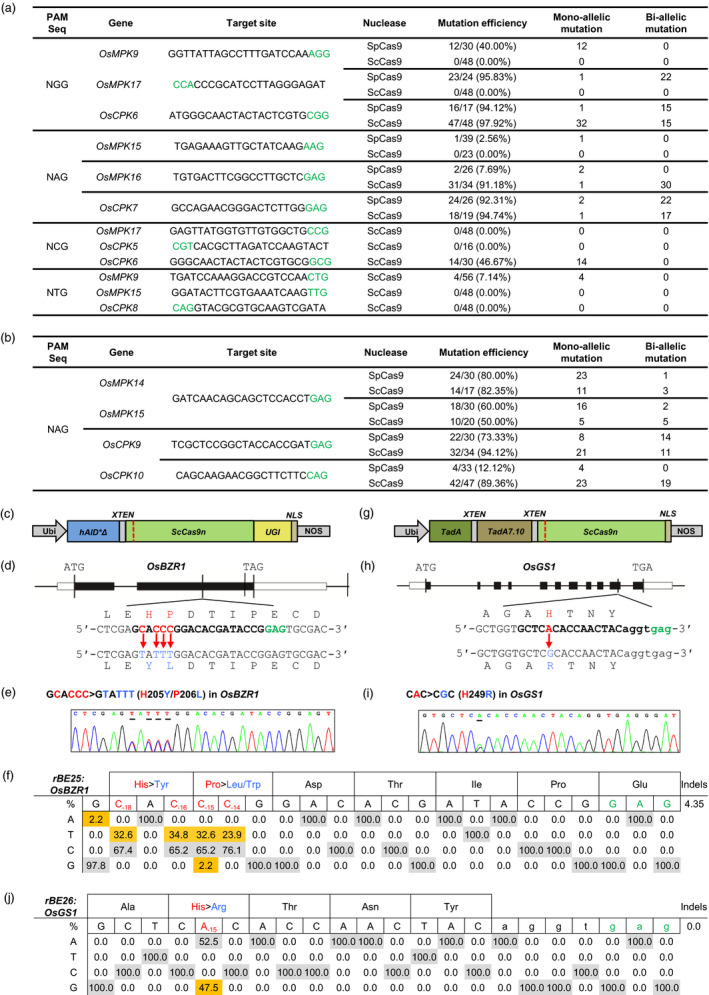
Targeted genome editing in rice using ScCas9, rBE25 and rBE26. (a) Summary of the frequencies of mutations induced by SpCas9 and ScCas9 towards different PAM sequences in T0 transgenic rice plants. (b) Summary of the simultaneous editing frequencies of the target genes induced by SpCas9 and ScCas9 towards NAG PAM in T0 transgenic rice plants. (c) Gene construct of cytidine base editor rBE25. (d) The target site in the OsBZR1 gene in rice. (e) Representative Sanger sequencing chromatogram of the rBE25‐edited OsBZR1 allele in a T0 transgenic line. (f) Summary of nucleotide changes in the editing window of the endogenous OsBZR1 gene caused by rBE25 in independent T0 transgenic lines. (g) Gene constructs of adenine base editor rBE26. (h) The target site in the OsGS1 gene in rice. (i) Representative Sanger sequencing chromatogram of the rBE26‐edited OsGS1 allele in a T0 transgenic line. (j) Summary of nucleotide changes in the editing window of the endogenous OsGS1 gene caused by rBE26 in independent T0 transgenic lines. The intron is depicted as lower‐case letters. The PAM sequences, target sequences, candidate bases in the putative editing window and detected nucleotide changes/corresponding amino acids are highlighted in green, bold, red and blue, respectively. In (e) and (i), the nucleotide changes are underlined in the sequencing chromatograms
