Figure 1.
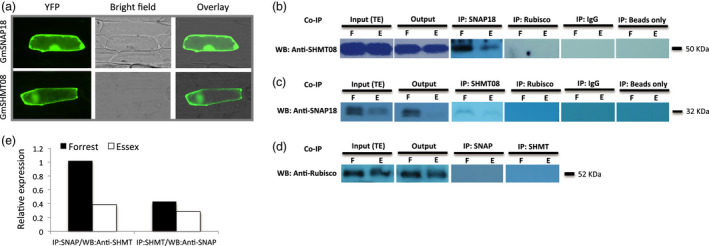
Subcellular localization and interaction analyses of GmSNAP18 and GmSHMT08 proteins by co‐immunoprecipitation (Co‐IP). (a) The coding sequences of GmSNAP18 and GmSHMT08 (used as positive control) were fused to the N‐terminal end of the eYFP and delivered into onion epidermal cells using biolistic bombardment. YFP fluorescence was localized in the cytoplasm and plasma membrane for GmSNAP18 like GmSHMT08. (b) IP: The total protein extracts of soybeans Forrest and Essex from SCN‐infected roots were immunoprecipitated with anti‐SNAP18 polyclonal antibodies (PA) or anti‐Rubisco PA (used as negative control). Blots from the eluted fraction were probed with anti‐SHMT08 PA or anti‐Rubisco PA. (c) The total protein extracts of roots of soybeans Forrest and Essex were immunoprecipitated with anti‐SHMT08 PA or anti‐Rubisco PA (as the control). Blots from the eluted fraction were probed with anti‐SNAP18 PA or anti‐Rubisco PA. Total protein extracts from input (TE) and output were blotted as well. (d) No band observed when using anti‐Rubisco as a negative control from both Co‐IP:SHMT08 and Co‐IP:SNAP18. Only IgG or beads were used for Co‐IP experiments as a negative control and technical control, respectively. The Co‐IP results indicated that GmSNAP18 interacts physically with GmSHMT08 and vice versa in two independent pull‐down experiments. (e) Relative expression of the GmSHMT08 and GmSNAP18 interaction intensity between Forrest and Essex from SDS‐PAGE was measured using ImageJ software and normalized using the Rubisco expression as reference. WB, Western blot.
