Figure 4.
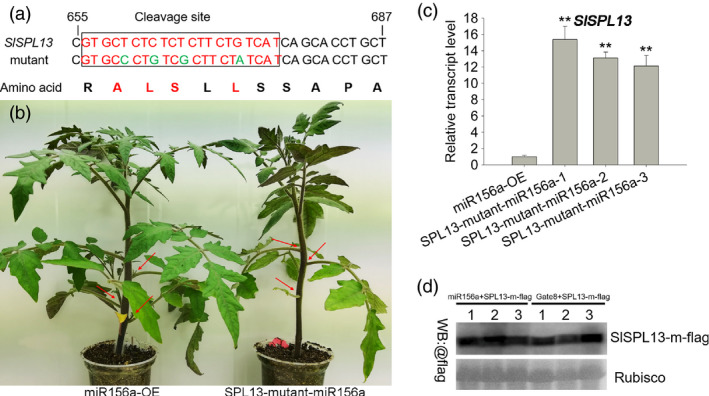
Phenotypic characterization of plants overexpressing a site‐directed mutant allele of SPL13 (pCAMBIA 1302, hygromycin resistance) in the 35S‐miR156a background. (a) Four nucleotides were changed in the cleavage site of the SPL13. These mutations prevent cleavage but do not change the amino acid sequence. This mutant allele of SPL13 was amplified using nested PCR. (b) Lateral branch phenotype in the SPL13 mutant and miR156a lines. Red arrows indicate the leaf axils. (c) Quantitative PCR analysis of SPL13 expression in young leaves from three transgenic and miR156a lines. Asterisks indicate statistically significant differences relative to the miR156a‐OE and were determined using t‐tests. **, P < 0.01. (d) FLAG‐tagged SPL13 mutant and 35S‐miR156a constructs transiently co‐expressed in the leaves of N. benthamiana. The FLAG‐tagged SPL13 mutant (SPL13‐m‐flag) and the empty vector (pHELLSGATE8) were transiently co‐expressed. Leaf extracts were analysed by Western blotting (WB) using anti‐FLAG antibodies. These experiments were repeated three times and yielded similar results each time.
