Fig. 3. Restoring the Balance of H3K27 Methyltransferase Activity to H3K27 Demethylase Activity Rescues Myogenic Differentiation Under Hypoxic Conditions.
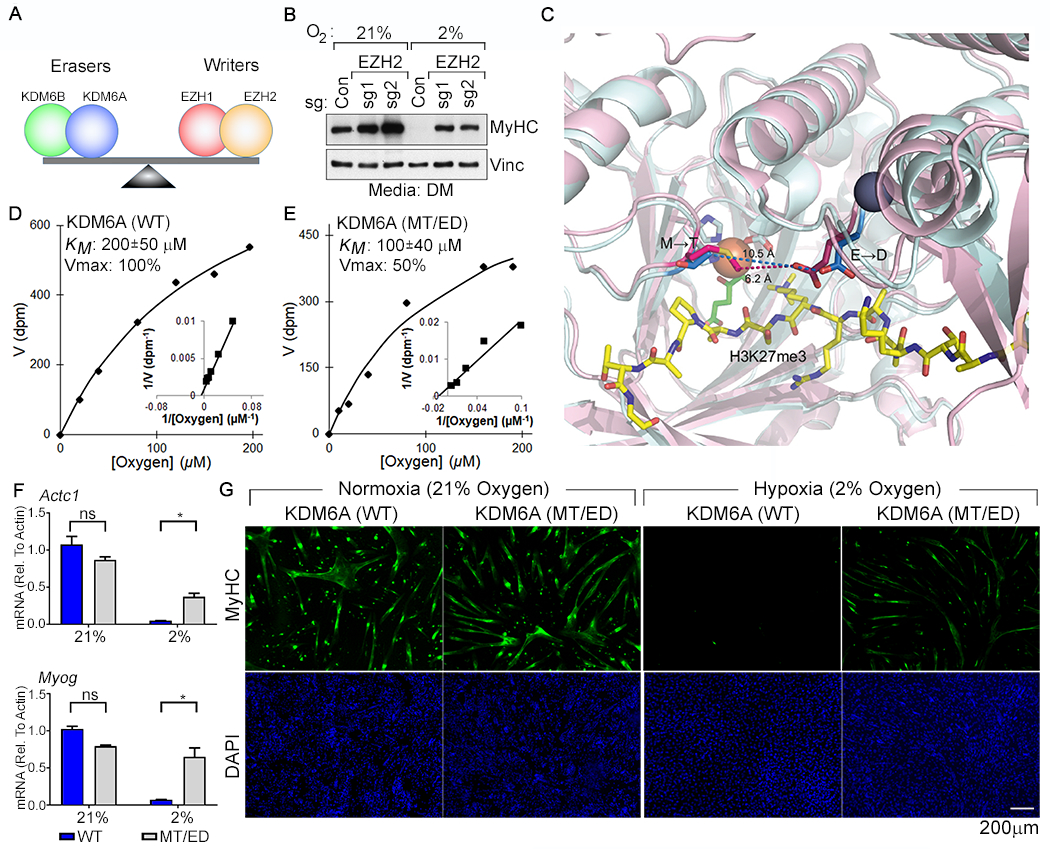
(A) Model for control of H3K27 methylation by the indicated opposing demethylases (“erasers”) and methyltransferases (“writers”). (B) Immunoblot analysis of C2C12 cells lentivirally transduced to express the indicated sgRNAs and cultured under the indicated conditions. (C) Structural models of the KDM6A (pink) and KDM6B (cyan) catalytic pockets. The non-conserved M1190 (KDM6A) → T1434 (KDM6B) and the E1335 (KDM6A) → D1579 (KDM6B) are highlighted. Peptidic H3K27me3 substrate (yellow), Fe+2 (orange), 2-oxoglutarate (green), and Zn+2 (grey) are shown. (D-E) Michaelis-Menten plots (inset Lineweaver-Burk plot) and measured oxygen KM and Vmax values (mean +/− SD, N=3) of recombinant KDM6A wild-type and the MT/ED mutant. (F-G) Real-Time qPCR analysis (mean +/− SD, N =3) of the indicated genes (F) and immunofluorescence analysis (G) of C2C12 cells transduced to produce wild-type human KDM6A or the KDM6A MT/ED variant and then cultured in DM at the indicated oxygen concentrations for 4 days.
