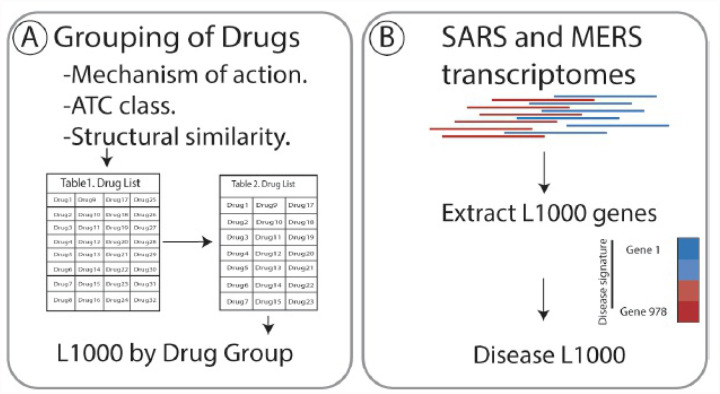
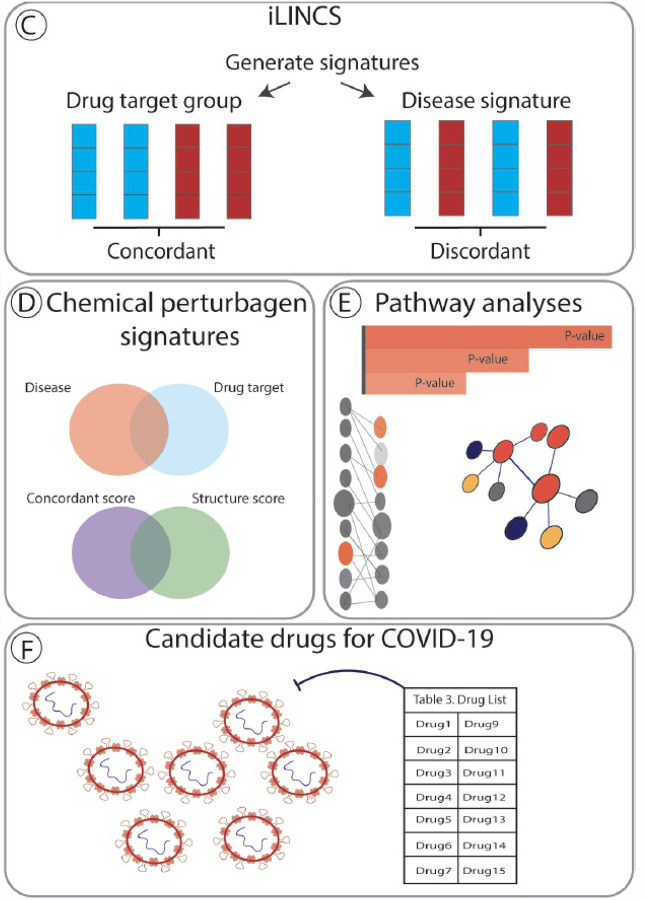
Figure 1.
Overview of the workflow to identify candidate repurposable drugs to combat COVID-19. A) Drugs that are currently in use to treat coronavirus and putative COVID-19 treatments were clustered based on mechanism of action, ATC class and structural similarity. B) Gene expression data of the 978 genes that comprise the Library of Integrated Network-Based Cellular Signature (iLINCS) L1000 genes were extracted from severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome- related coronavirus (MERS) (GSE56192) transcriptomic datasets. C) Consensus iLINCS gene signatures were generated for drug groupings and disease. D) Connectivity analysis was conducted and a list of chemical perturbagens that are concordant (≥ 0.321 concordance) to the drug target grouping signatures or discordant (≤ − 0.321 discordance) to the disease signatures was generated. Chemical perturbagens are curated to identify top candidate drugs. E) Biological pathways of drug target groupings, disease signatures and candidate drugs was conducted. F) Fourteen perturbagens with concordance scores ≥ 0.8 in MCF7 and HA1E cell lines were shortlisted as repurposable candidate drugs.