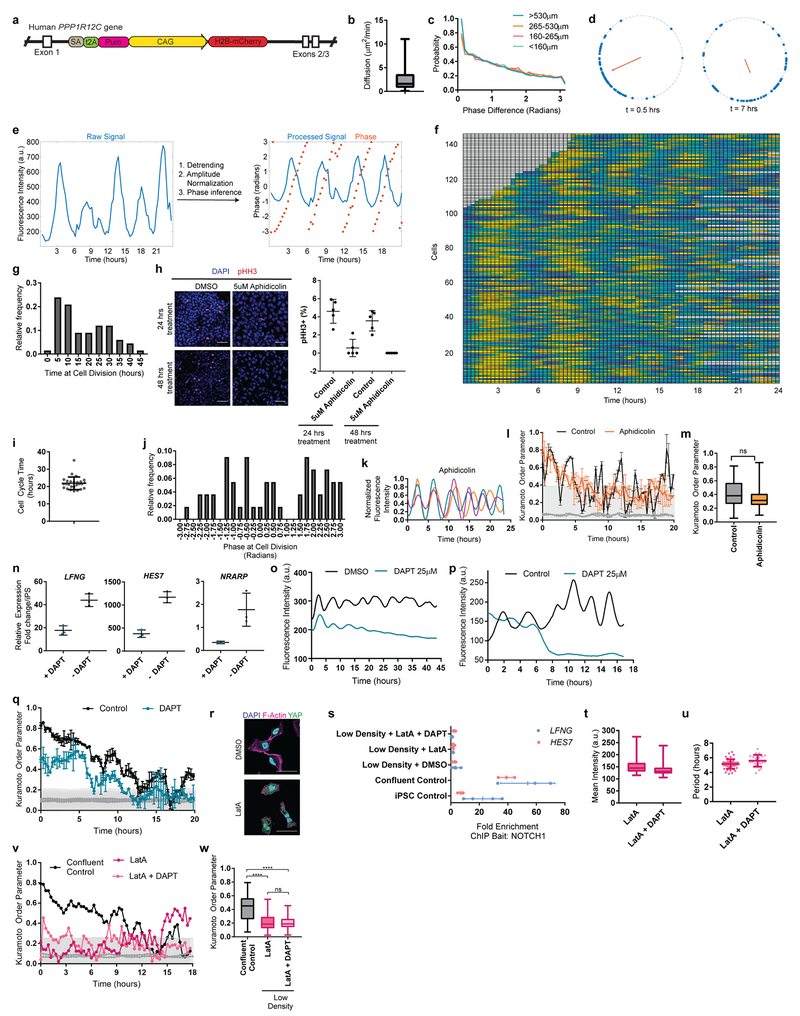
Extended Data Figure 5.
a, Scheme showing the insertion of a constitutively expressed pCAG-H2B-mCherry nuclear label in the safe harbor AAVS1 locus in a HES7-Achilles human iPSC background. b, Diffusion (μm2/min) for individual human HES7-Achilles cells automatically tracked over a period of 24 hours. Middle hinge corresponds to median, lower and upper hinges correspond to 1st and 3rd quartiles, lower and upper whiskers correspond to minimum and maximum. n=76 c, Distribution of pairwise instantaneous phase shifts between individual oscillating human HES7-Achilles cells, binned by instantaneous distance between pairs of cells. P-values for the pair-wise Kolmogorov-Smirnov test are as follows: 0.6407, 0.1811, 0.1340, 0.1428, 0.6784, 0.8171. n=1000. d, Distribution of phases along the unit circle at early, middle, and late timepoints. Each dot represents one cell. n=144 cells. e, Illustration of phase determination: representative raw HES7-Achilles fluorescence profile for an automatically tracked cell (left) and corresponding processed signal along with the inferred phase from Hilbert transform (right). f, Heatmap of HES7-Achilles fluorescence intensity over time in automatically tracked cells. Each line represents one cell. n=144 cells. g, Histogram of the time (hours since the onset of imaging) at cell division for manually tracked human HES7-Achilles cells. n=67 h, Left: Immunofluorescence staining for phosphorylated histone H3 (Ser10) in human iPSC-derived PSM cells treated with either vehicle control (DMSO) or 5μM Aphidicolin for 24 or 48 hours, starting on day 2 of differentiation. n=5. Scale bar = 100μm. Right: Quantification of phosphorylated histone H3 (Ser10) nuclei as a percent of total nuclei. Middle hinge corresponds to median, lower and upper hinges correspond to 1st and 3rd quartiles, lower and upper whiskers correspond to minimum and maximum. i, Scatter plot showing the cell cycle time in human iPSC-derived PSM cells cultured in CLFBR medium. Mean±SD. n=26. j, Scatter plot showing the cell cycle time in human iPSC-derived PSM cells cultured in CLFBR medium. Mean±SD. n=26. k, Normalized HES7-Achilles fluorescence intensity profiles for 3 individual human iPSC-derived PSM cells pre-treated with 5μM Aphidicolin for 24 hours. n=6 independent experiments. l, Kuramoto order parameter over 20 hours on day 3 of differentiation for human HES7-Achilles cells treated with vehicle control (DMSO) or 5μM Aphidicolin for 24 hours. Synchronization threshold is shown as the mean±SD of the Kuramoto order parameter for same dataset, but with randomized phases. n=45 cells (Control), 48 cells (Aphi). m, Comparison of the Kuramoto order parameter for oscillating HES7-Achilles treated with vehicle control (DMSO) or 5μM Aphidicolin. Middle hinge corresponds to median, lower and upper hinges correspond to 1st and 3rd quartiles, lower and upper whiskers correspond to minimum and maximum. Paired two-sided t-test p=0.348. n=45 cells (Control), 48 cells (Aphi). n, qRT-PCR for Notch target genes HES7, NRARP and LFNG in human iPSC-derived PSM cells treated with vehicle control (DMSO) or 25μM DAPT on day 2 of differentiation. Mean±SD. n=3. o, Example of HES7-Achilles fluorescence intensity in a small region of interest (ROI) over a period of 45 hours in cells treated with DMSO (vehicle control) or the γ-secretase inhibitor DAPT (25μM) in CLFBR medium. n=16 independent experiments p, Representative example of Hes7-Achilles fluorescence intensity profiles for mESC-derived PSM cells treated with vehicle control (DMSO) or 25μM DAPT. n=13 independent experiments q, Kuramoto order parameter over 20 hours on day 2 of differentiation for human HES7-Achilles cells treated with vehicle control (DMSO) or 25μM DAPT. Synchronization threshold is shown as the mean±SD of the Kuramoto order parameter for same dataset, but with randomized phases. n=131 cells (Control), 110 cells (DAPT). r, Representative immunofluorescence staining for YAP, F-actin (phalloidin) and DAPI nuclear stain in isolated human PSM-like cells treated with either DMSO or Latrunculin A (350nM). Scale bar = 50 μm. n=4 independent experiments. s, ChIP-qPCR fold enrichment of the LFNG and HES7 promoters in chromatin pulled down with an antibody against NOTCH1 relative to isotype IgG controls. Mean ±SD. iPSC control n=4, all other conditions n=3. t, Mean HES7-Achilles fluorescence intensity for isolated human cells cultured with either 350nM Latrunculin A alone or in combination with 25μM DAPT. Middle hinge corresponds to median, lower and upper hinges correspond to 1st and 3rd quartiles, lower and upper whiskers correspond to minimum and maximum. n=18 cells. u, Scatter plot showing the HES7-Achilles oscillatory period for isolated human cells cultured with either 350nM Latrunculin A alone or in combination with 25μM DAPT. Mean ±SD. n= 47 (LatA), 22 (LatA + DAPT) v, Kuramoto order parameter over 18 hours on day 2 of differentiation for human HES7-Achilles cells treated with DMSO, LatA alone or LatA in combination with DAPT. Synchronization threshold is shown as the mean±SD of the Kuramoto order parameter for same dataset, but with randomized phases. n=53 cells (Control), 18 cells (LatA), 18 cells (LatA + DAPT). w, Comparison of the Kuramoto order parameter in confluent HES7-Achilles cells vs. isolated cells treated with either 350nM Latrunculin A alone or in combination with 25μM DAPT. Middle hinge corresponds to median, lower and upper hinges correspond to 1st and 3rd quartiles, lower and upper whiskers correspond to minimum and maximum. Paired one-way ANOVA with Bonferroni correction: Confluent control vs. LatA p=1.16e−6, Confluent control vs. LatA + DAPT p=6.8e−13, LatA vs. LatA + DAPT p=0.304. n=53 cells (Control), 18 cells (LatA), 18 cells (LatA + DAPT).