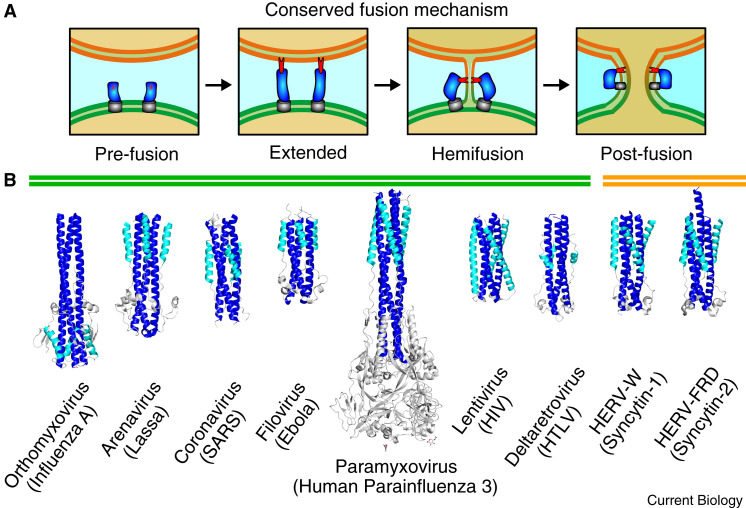
Figure 1.
Conserved mechanism for membrane fusion and the class I fusogen superfamily
(A) Viral fusion proteins (blue) are displayed in a pre-fusion state, with their carboxy-terminal transmembrane region (grey) attached to the virion membrane (green), and their hydrophobic fusion peptide or fusion loop (red) safely buried. The protein then goes through a series of conformational changes, extending to embed its fusion peptide/loop in the host cell membrane (orange) before bringing the carboxy-terminal transmembrane region and fusion peptide/loop together in a post-fusion state. (B) Structures of the viral class I fusogens from major human pathogens are presented, including influenza A (PDB: 1HTM), Lassa virus (PDB: 5OMI), SARS-CoV (PDB: 1WNC), Ebola (PDB: 2EBO), human parainfluenza virus-3 (PDB: 1ZTM), HIV-1 (PDB: 3WFV), human T-lymphotropic virus-1 (PDB: 1MG1), as well as the two solved structures for human syncytin-1 (PDB: 6RX1) and syncytin-2 (PDB: 6RX3). The amino-terminal helix (heptad repeat-1) is coloured blue, the carboxy-terminal helix (heptad repeat-2) is coloured cyan, and the connecting chain reversal region (if present) is coloured gray. The structures shown are in their post-fusion trimeric states.