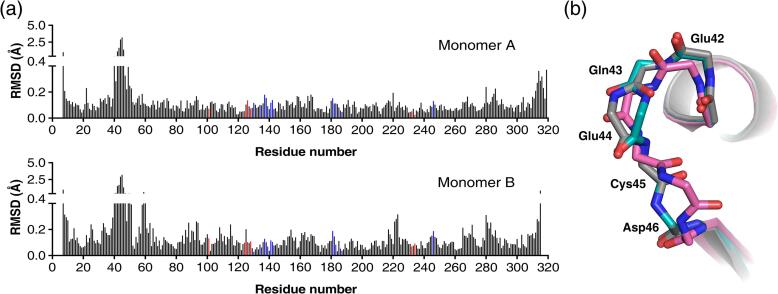
Fig. 5.
Comparison of the overall structure of the Arginase-1/ABH complexes at pH 7.0 and 9.0. (a) Root-mean-square deviation (RMSD) between the main chain atoms (Cα, C, O and N) of the pH 7.0 complex (PDB ID: 6Q92) and the pH 9.0 complex (PDB ID: 6Q9P) calculated per residue. Red bars indicate manganese-coordinating residues, while blue bars indicate active site residues interacting with ABH. (b) Superposition of the backbone atoms of the Glu42 to Asp46 surface loop in the two structures (pH 7.0 in cyan and pH 9.0 in magenta) and the previously reported Arginase-1/ABH crystal structure (PDB ID: 2AEB; grey).