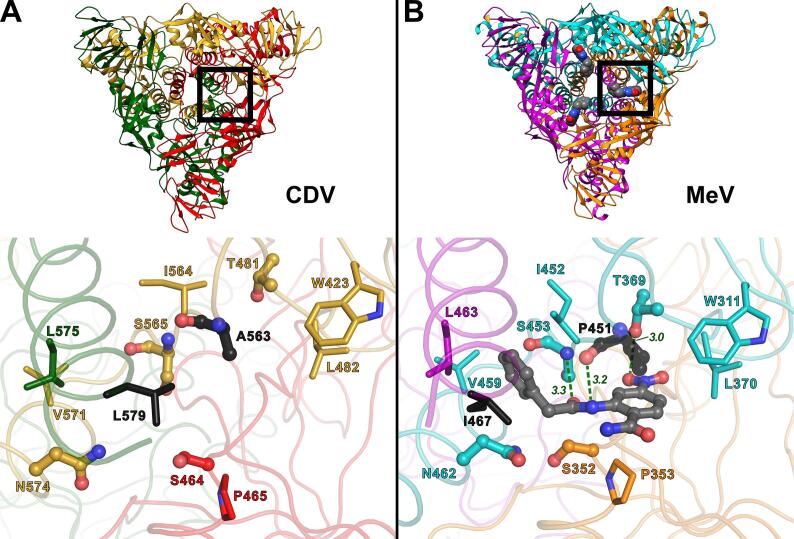
Fig. 4.
Comparison of the inhibitor binding pocket of CDV solF and MeV solF. Bottom views of the prefusion CDV solF (A) or MeV solF (PDB entry 5YZC; (Hashiguchi et al., 2018)) structures (B) are displayed (top). In the models, the three protomers are differently colored: (A) yellow, red and green; (B) cyan, orange and magenta. A magnified view of the CDV solF binding pocket (A) and the same view of the MeV solF binding pocket with bound AS-48 (B) are shown (bottom). The side chains of the amino acids within a distance of 4 Å from the AS-48 molecule in MeV solF and the corresponding amino acids in CDV solF are displayed. The hydrophobic amino acid residues are displayed as sticks and the amino acid residues in hydrogen bonding distance to AS-48 in MeV solF (B) (and the corresponding amino acids in CDV solF (A)) are displayed as ball-and-sticks. The hydrogen bonds to AS-48 are indicated with broken lines and the distances are given in Å. Amino acids that are different between CDV and MeV are colored in black. The protomers are colored as in the models in the top panel.