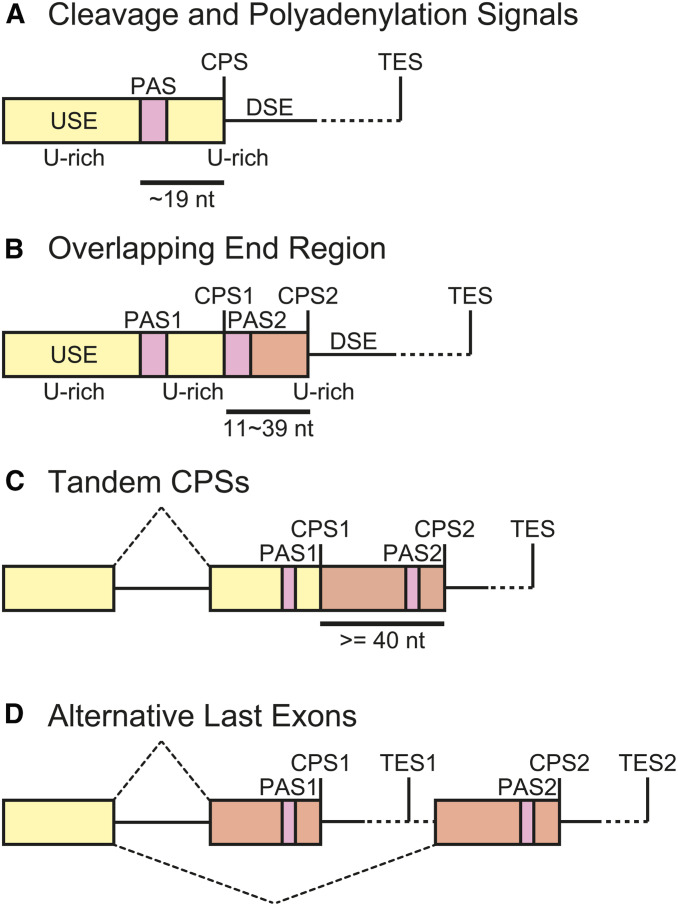
Figure 8.
Schematic representations of cis-elements for 3′ end processing of mRNAs in C. elegans. (A) Typical cleavage and polyadenylation signals. AAUAAA or related sequences embedded in a U-rich region functions as a polyadenylation signal (PAS). CPS, cleavage and polyadenylation site; DSE, downstream element; TES, transcription end site; USE, upstream element. (B) Overlapping end region (ORE). Two closely located (11–39 nt apart) CPSs have their own PAS and share a U-rich region as either DSE or USE. (C) Tandem cleavage and polyadenylation sites (CPSs). Two or more PASs and CPSs are located in the same last exon but are ≥ 40 nt apart. (D) Alternative last exons (ALEs). Choice of the CPS is coupled with choice of the last exon of an mRNA. Boxes represent exons and solid lines indicate introns or 3′ flanking regions.