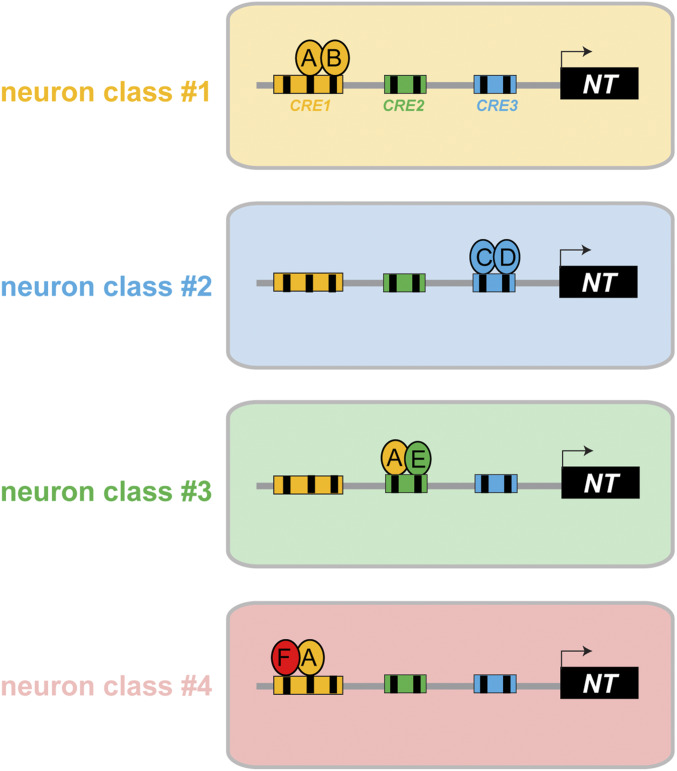
Figure 9.
Illustrations of the organization of cis-regulatory elements for neuron type-specific gene expression. A neurotransmitter pathway gene (NT) is controlled by different sets of transcription factors (A–F) in four different neuron classes. Reporter gene analysis (“promoter bashing”) has revealed distinct cis-regulatory elements in the NT locus that drive expression in four different neuron classes. The indicated transcription factors act through specific binding sites (black boxes) within a given cis-regulatory element. The following scenarios are illustrated: comparing neuron class 1 and class 2 illustrates the simplest scenario of modularity where different combinations of neuron type-specific transcription factors (A + B or C + D) control NT expression in different neuron classes. Comparing neuron class 1 and 3 illustrates that the same transcription factor A can be used in different neuron types, but it operates through distinct, separable cis-regulatory elements that each contain a cognate binding site for this transcription factor. One example for this are the two distinct elements from the unc-17/VAChT locus that drive expression in distinct sets of UNC-86-dependent neurons (RIH vs. IL2/URA/URB), with each of these elements containing validated UNC-86-binding sites (see Results). Comparing neuron class 1 and 4 illustrates a cis-regulatory element that drives expression in multiple distinct neuron classes, and for both neuron classes the same transcription factor-binding site and the same cognate transcription factor are required (e.g., unc-17 element driving UNC-86-dependent expression in IL2, URA, and URB). In such a case, UNC-86 may interact with different transcription factors in neuron 1 vs. neuron 4, but it will not be possible to generate reporters that separate expression in neuron 1 and neuron 4. Hence, the modularity of cis-regulatory control is not manifested by distinct separable elements, but by distinct transcription factor-binding sites that are used in a modular and combinatorial manner.