Extended Data Fig. 9: RMMAI accumulation and low-level DSB formation on mo-2 regions in oocytes.
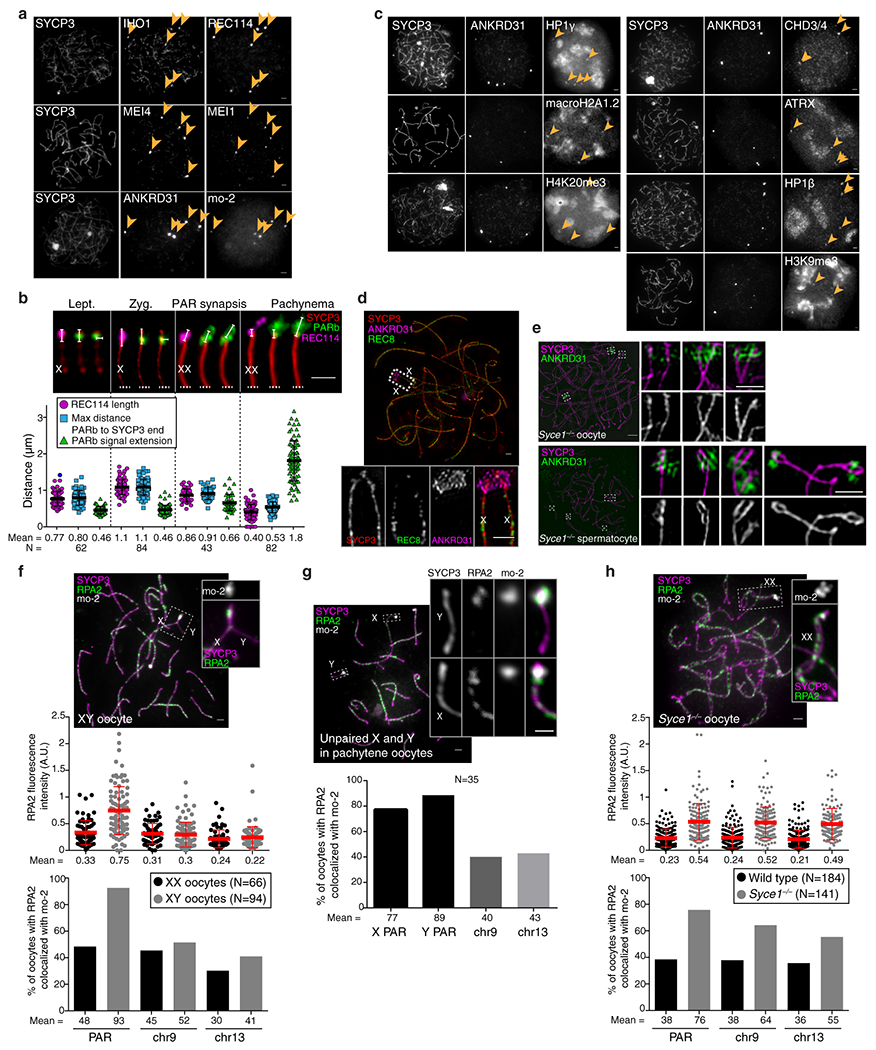
(a) Examples of zygotene oocytes showing the colocalization between blobs of IHO1 and REC114, MEI4 and MEI1, or ANKRD31 and mo-2 FISH signal (arrowheads). Scale bars: 2μm. RMMAI blobs colocalized with mo-2 FISH signals in all zygotene oocytes analyzed (N>30) from at least three mice. (b) PAR ultrastructure in oocytes, quantified as in Extended Data Fig. 3b. Late zygotene cells with PAR synapsis are compiled separately from other zygotene cells. Error bars: means ± SD. Scale bar: 1 μm. (c) Examples of zygotene oocytes showing colocalization of ANKRD31 blobs with enrichment for heterochromatin factors. Scale bars: 2 μm. ANKRD31 colocalized with heterochromatin factors blobs in all zygotene oocytes analyzed (N>20) from one mouse. (d) Representative SIM image of a wild-type late zygotene oocyte showing neither detectable splitting of the PAR axis nor REC8 enrichment. Scale bar: 2 μm. The absence of spermatocyte-like differentiation of the PAR axis was observed (N>30 zygotene oocytes) in more than three mice. A modest degree of differentiation was observed in a minority of oocytes (5/45) analyzed by SIM, but this did not resemble the typical PAR axis splitting found in spermatocytes. (e) Prolonged asynapsis does not allow axis splitting to occur in oocytes. Because synapsis appears sufficient to trigger collapse of PAR ultrastructure in spermatocytes (Extended Data Fig. 3b), we asked if preventing synapsis (i.e., in a Syce1−/− mutant) could reveal a cryptic tendency toward axis splitting in oocytes. However, whereas axis splitting was clearly observed by SIM in Syce1−/− mutant spermatocytes, PAR axes were not detectably split in oocytes. Scale bars: 2 μm for main micrograph, 1 μm for insets. Axis splitting of chr9 was observed by SIM in multiple (N>20) Syce1−/− spermatocytes from three different mice. The chr13 or chr4 centromere-distal axes were also occasionally seen to be split, but we did not quantify this for these chromosomes. In males, the differentiation of the PAR or the chr9 axes becomes hardly detectable at later stages in some pachytene-like spermatocytes as cells enter apoptosis, similar to Spo11−/− or Hormad1−/− mice. However, in Syce1−/− oocytes, no significant axis differentiation or splitting was observed by conventional microscopy or by SIM in multiple spermatocytes (N>30) from three different mice, similar to what we observed in wild-type oocytes. (f,h) Delaying synapsis promotes PAR DSB formation in oocytes. Top panels: representative micrographs of pachytene XY (f) and Syce1−/− XX oocytes (h). Middle panels: RPA2 fluorescence intensity at the border of mo-2 FISH signals from PAR, chr9, and chr13. Bottom panels: Percentage of oocytes with RPA2 focus colocalizing with mo-2 regions on PAR, chr9, and chr13. Graphs show data only for pachytene oocytes in which PARs are synapsed (two mice of each genotype). Error bars: means ± SD. Scale bars: 2 μm. (g) Percentage of pachytene oocytes with one or more RPA2 foci colocalizing with mo-2 FISH signal from PAR, chr9 and chr13 in XY pachytene oocytes that had unsynapsed X and Y chromosomes. Scale bar: 2 μm, inset: 1 μm.
