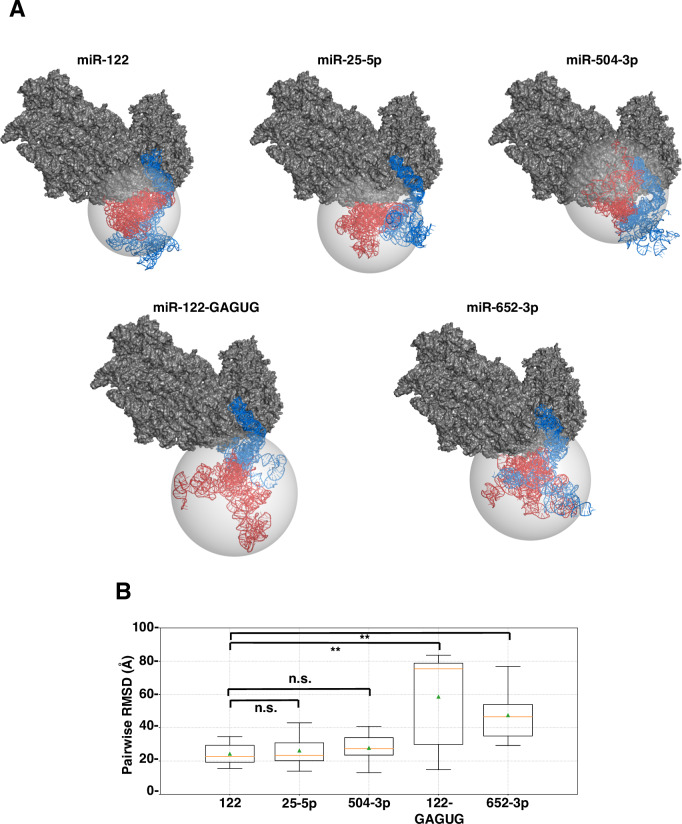
Fig 5. Model of miRNA:HCV-RNA interaction.
(A) Superposition results of HCV-RNA tertiary structure onto a Cryo-EM reference structure of the HCV IRES bound to the human ribosome. Representative models of each miRNA (top) were displayed. miRNA binding regions and long arm composed of SLII (nt1-117: blue) and SLIIIa-IIId (nt118-333: red) of HCV-RNA, miRNAs (blue) and 40S rRNA subunit (gray) were shown. Representative models of each miRNA:HCV-RNA:Ago2 (bottom). The distribution of tertiary structures of SLIIIa-IIId were shown as gray sphere. (B) Box-plots of pairwise RMSDs among models of HCV region (SLIIIa-IIId: nt118-333, red blocks in Fig 5A, top). All pairwise RMSDs among 5 representative models (Fig 5A) of nt118-333 regions were determined. The bottom and top boundaries of the box correspond to Q1 (25th percentile) and Q3 (75th Percentile) quartiles, respectively. The lower and upper whiskers correspond to Q1–1.5 * IQR and Q3 + 1.5 * IQR, respectively, where IQR = Q3 –Q1. Median value (Q2, 50th percentile) is indicated by an orange line within the box, and mean value is indicated by a triangle symbol in green. Asterisks indicate significant differences (**P < 0.01, n.s.: not significant).