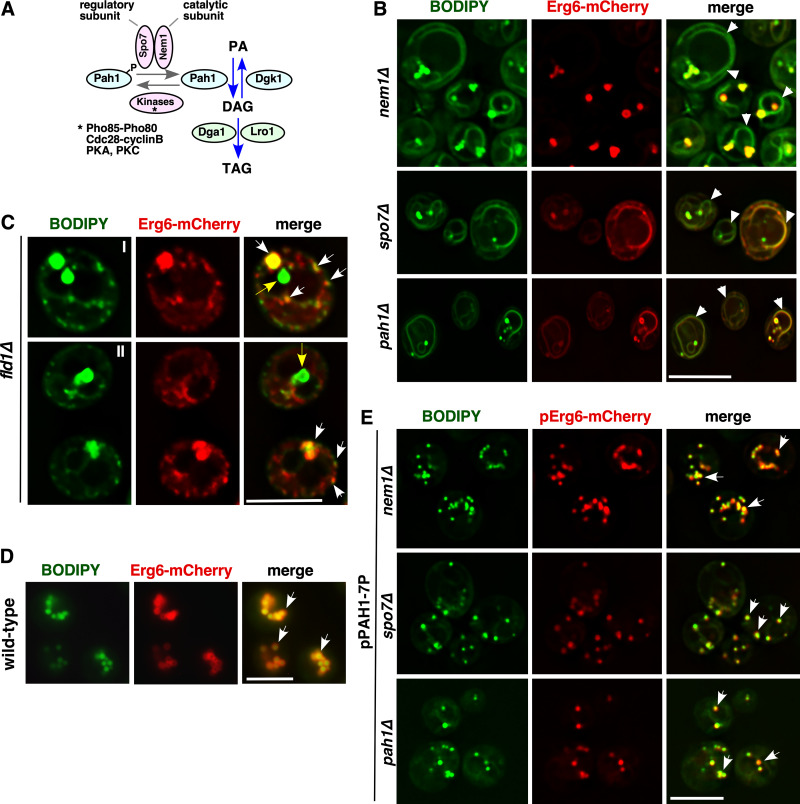
Figure S2.
Data associated with Fig. 2. Cells lacking Nem1, Spo7, Pah1, or Fld1 accumulate neutral lipids in the ER and have aberrant droplets. (A) Diagram depicting the regulation of triglyceride synthesis. The Nem1-Spo7 complex dephosphorylates and thereby activates the phosphatidate phosphatase Pah1, resulting in formation of DAG from phosphatidic acid (PA). DAG then serves as a substrate for the TAG-synthesizing enzymes Dga1 and Lro1. (B) BODIPY accumulates in the ER of nem1Δ, spo7Δ, and pah1Δ cells. Yeast cells of the indicated genotype expressing genomic Erg6-mCherry were stained with BODIPY. White arrowheads denote BODIPY accumulation in the ER membrane. Scale bar, 5 µm. (C) Fld1-deleted cells accumulate numerous small and a few supersized LDs. fld1Δ cells expressing genomic Erg6-mCherry were stained with BODIPY. White arrows denote colocalization between BODIPY and Erg6-mCherry. Yellow arrows denote BODIPY-stained supersized puntae that do not colocalize with Erg6-mCherry. Two examples are shown (I, II). Scale bar, 5 µm. (D) WT cells do not show BODIPY accumulation in the ER. BODIPY staining of WT cells expressing genomic Erg6-mCherry. White arrows denote colocalization between BODIPY and Erg6-mCherry. Scale bar, 5 µm. (E) Pah1-7P rescues the LD biogenesis defect of nem1Δ, spo7Δ, and pah1Δ mutant cells. BODIPY staining of nem1Δ, spo7Δ, and pah1Δ mutants expressing Erg6-mCherry and Pah1-7P. White arrows denote colocalization between BODIPY and Erg6-mCherry. Scale bar, 5 µm.