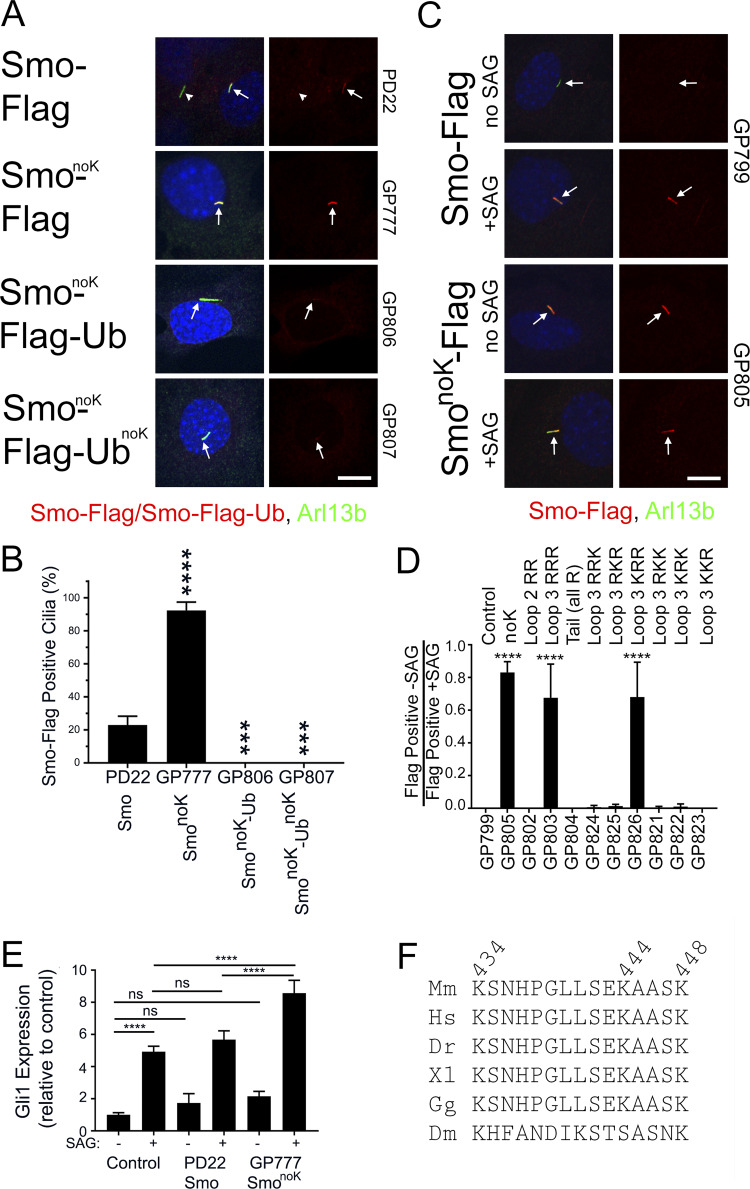
Figure 5.
Smo loop three lysines are required for regulated ciliary localization. (A) Wild-type MEFs transfected with Smo-Flag (PD22), SmonoK-Flag (GP777), SmonoK-Flag-Ub (GP806), or SmonoK–Flag-UbnoK (GP807) were serum starved, fixed, and stained for Smo (Flag; red), cilia (Arl13b; green; arrows), and nuclei (DAPI; blue). Scale bar is 10 μm and applies to all images. Each image is maximum projection of a three-image stack taken at 0.2-μm intervals. Left image of each pair is a three-color composite; right image shows only the red Flag channel. (B) Presence of ciliary Smo-Flag or Smo-Flag-Ub was quantitated from cells in A. N is 3 replicates with at least 100 cells counted per condition. ****, P < 0.0001; ***, P < 0.001 compared with control by one-way ANOVA. Error bars indicate SD. (C) Wild-type MEFs transfected with GgCryD1-Bsd-IRES-Smo-Flag (GP799), GgCryD1-Bsd-IRES-SmonoK-Flag (GP805) were serum starved, treated ±SAG, fixed, and stained for Smo (Flag; red), cilia (Arl13b; green; arrows), and nuclei (DAPI; blue). Scale bar is 10 μm and applies to all images. Each image is maximum projection of a three-image stack taken at 0.2-μm intervals. Left image of each pair is a three-color composite; right image shows only the red Flag channel. (D) Ciliary Smo was quantitated from the cells described in C as well as a series of mutants designed to identify the lysines important for regulated localization of Smo to cilia. Results are reported as a ratio of the percentage of cells with Smo-positive cilia without SAG divided by the percentage of cells with Smo-positive cilia after treatment with SAG. N is 3 replicates with at least 100 cells counted per condition. ****, P < 0.0001 compared with control (GP799) by one-way ANOVA. Error bars indicate SD. (E) Wild-type MEFs untransfected (control) or transfected with Smo-Flag (PD22) or SmonoK-Flag (GP777) were serum starved and treated ±SAG for 24 h. RNA was collected, reverse transcribed, and quantitated using real-time PCR. N is 3 replicates. Data are reported with respect to wild-type unstimulated (−SAG). ****, P < 0.0001; ns, not significant by two-way ANOVA. Error bars indicate SD. (F) Sequence from Smo cytoplasmic loop three showing complete conservation in vertebrate models. Drosophila melanogaster (Dm) retained three lysines but was otherwise divergent. Dr, Danio rerio; Hs, Homo sapiens; Gg, Gallus gallus; Mm, Mus musculus; Xl, Xenopus laevis.