Cells must control the composition and location of biomolecular condensates. This study shows that phosphorylation of the RNA-binding protein Whi3 is used to regulate functionally distinct condensates important for cell polarity and nuclear division in multinucleate fungus Ashbya gossypii.
Abstract
Biomolecular condensation is a way of organizing cytosol in which proteins and nucleic acids coassemble into compartments. In the multinucleate filamentous fungus Ashbya gossypii, the RNA-binding protein Whi3 regulates the cell cycle and cell polarity through forming macromolecular structures that behave like condensates. Whi3 has distinct spatial localizations and mRNA targets, making it a powerful model for how, when, and where specific identities are established for condensates. We identified residues on Whi3 that are differentially phosphorylated under specific conditions and generated mutants that ablate this regulation. This yielded separation of function alleles that were functional for either cell polarity or nuclear cycling but not both. This study shows that phosphorylation of individual residues on molecules in biomolecular condensates can provide specificity that gives rise to distinct functional identities in the same cell.
Introduction
A central feature of cell organization is the assembly and regulation of compartments. Cellular biochemistry is spatially limited by sequestration into both membrane-bound organelles and membraneless bodies termed biomolecular condensates (Banani et al., 2017). While there are many condensates in both the cytosol and nucleus, mechanisms that control where and when they form, determine their physical properties, and establish their distinct functions remain under study (Brangwynne et al., 2011; Banani et al., 2017). Physical principles for phase transitions indicate that condensates should be sensitive to local changes in the abundance of key components, changes in the solvent conditions, or electrostatic interactions among components.
Work on the RNA-binding protein Whi3 in the filamentous fungus Ashbya gossypii has led to some key insights into how specificity can arise. This protein forms assemblies dependent on an extended polyQ-tract and binding to specific mRNAs. CLN3 (a G1 cyclin) transcripts are heterogeneously positioned near nuclei in a Whi3-dependent manner. BNI1 (a formin) transcripts are positioned at growing tips and nascent polarity sites in a Whi3-dependent manner. The assembly of Whi3 structures is essential for asynchronous division of multiple nuclei in a common cytoplasm and the generation of new polarity sites (Lee et al., 2013, 2015). The fact that this single protein forms functionally distinct condensates in a shared cytoplasm makes it an excellent model to examine sources of specificity.
We hypothesized that localized signaling leads to specific post-translational modifications on Whi3 to control where and when it condenses and dissembles, or the function of a given complex. A. gossypii can grow to be hundreds of microns across with dozens of simultaneously elongating tips and nuclei that often travel long distances from their birthplace (Gladfelter et al., 2006; Schmitz et al., 2006). Given their large size and the potential to experience different environments in different parts of the cell, it could be advantageous for cells to use post-translational modifications controlled by local signaling to create a spatially restricted function to condensates.
The genetic tractability and clear phenotypic links to function for the Whi3 protein provide an opportunity to search for specificity at the level of post-translational modification of a protein that is a common component of different condensate assemblies. Remarkably, we identified clean separation of function mutants on two key sites on the protein that specifically block either the nuclear cycle or cell polarity functions. This indicates that despite being driven by low complexity sequences, there is exquisite sensitivity of condensates to even small changes in local charge.
Results and discussion
Identification of phosphorylation sites on Whi3
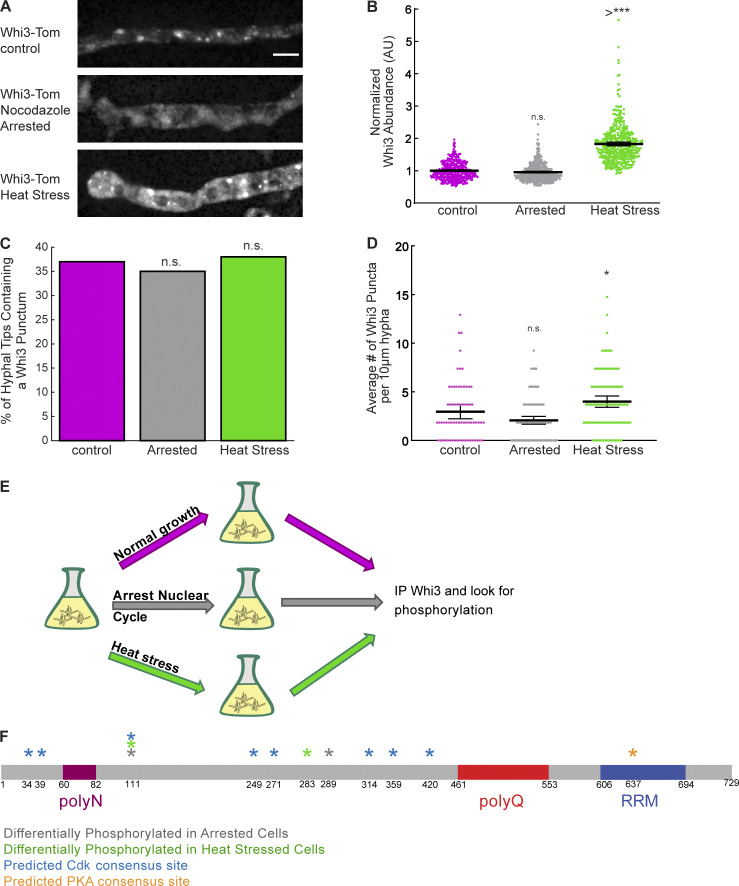
We hypothesized that the formation of Whi3 condensates is controlled at specific places and particular times in the nuclear division cycle. When cells are arrested in nocodazole, which leads to a G2/M arrest, we noticed that there was a modest decrease in the number of Whi3 puncta in the vicinity of nuclei, although this effect was not statistically significant (Fig. 1, A and D). In acute heat–shocked cells (2 h at 42°C), additional puncta formed in the hyphae, suggesting a redistribution of the protein into potentially distinct stress-induced condensates (Fig. 1, A and D). These conditions did not affect the number of hyphae containing Whi3 puncta at their tips (Fig. 1 C), and overall protein levels remained similar (Fig. 1 B). We hypothesized that rapid changes in the state of condensates could be triggered by post-translational modifications to Whi3 protein.
Figure 1.
Whi3 is differentially phosphorylated during nocodazole arrest and heat stress. (A) Representative images of Whi3-tomato protein localized in hyphae under different growth conditions. Scale bar, 5 µm. (B) Normalized Whi3 abundance as measured by fluorescence intensity of Whi3-tomato protein within the hypha under different growth conditions. n > 386 for all conditions. *, P < 0.05 by one-way ANOVA with multiple comparisons. Bars denote 95% confidence interval. ***, P < 0.0005. (C) Percent of hyphal tips containing a Whi3 punctum. n > 62 for all conditions. *, P < 0.05 by N-1 χ2 test with Benjamini–Hochberg correction. (D) Average number of Whi3 puncta per 5.410-µm hyphal segment. n > 66 for all conditions. *, P < 0.05 by one-way ANOVA with multiple comparisons. Bars denote 95% confidence interval. (E) Schematic of mass spectrometry workflow. (F) Diagram of identified or predicted phosphorylation sites. RRM, RNA-recognition motif; AU, arbitrary units; n.s., not significant
We therefore evaluated if Whi3 was differentially phosphorylated under cell cycle arrest or heat stress. Whi3-TAP was immunoprecipitated from A. gossypii cells growing asynchronously, arrested in mitosis, or under heat stress. The immunoprecipitate was analyzed by mass spectrometry to identify phosphorylation sites on Whi3 protein in each of the different conditions (Fig. 1 E). We found multiple (X = 22, Table 1) sites on the protein that were phosphorylated and several sites on the protein that were more phosphorylated in nuclear cycle arrest and heat stress conditions. The mass spectrometry did not fully cover the entire Whi3 protein, and repetitive sequences in the polyQ region as well as portions of the RNA-binding domain were not captured (Table 1). We replaced Whi3 at the endogenous locus under the native promoter with nonphosphorylatable and phosphomimetic versions of the protein tagged with TdTomato for each of the sites identified by the mass spectrometry as well as well as predicted Cdk and protein kinase A (PKA) sites (Mok et al., 2010; Mizunuma et al., 2013; Fig. 1 F). We then assayed these mutants for cell behaviors regulated by Whi3 condensation, nuclear asynchrony and polarized growth, and evaluated the localization of the mutant protein as well as the abundance of target mRNAs and the ability of mutant proteins to phase separate in vitro.
Table 1. Sites phosphorylated with any confidence.
| Asynchronous 1 | Arrested 1 | Heat Stress 1 | Asynchronous 2 | Arrested 2 | Heat Stress 2 | ||
|---|---|---|---|---|---|---|---|
| % Coverage | 51.30% | 52.30% | 7.10% | 70.90% | 73.90% | 75.70% | |
| S025 | C | ? | NC | C | NC | C | |
| S026 | X | X | NC | ? | NC | X | |
| T034 | Cdk-1 | X | X | NC | C | C | C |
| S039 | Cdk-2 | C | X | NC | C | C | X |
| S111 | Cdk-3 | X | X | NC | X | X | X |
| S198 | GIN4-3 | X | NC | NC | C | C | C |
| S249 | Cdk-4 | X | X | NC | X | X | X |
| S283 | GIN4-4 | X | X | X | X | C | X |
| S289 | C | X | NC | C | X | C | |
| S292 | ? | ? | NC | X | C | X | |
| T294 | YAK1-1 | X | C | NC | ? | C | C |
| T313 | ? | ? | C | C | C | X | |
| T314 | Cdk-6 (full) | ? | ? | C | ? | ? | ? |
| S355 | X | X | NC | X | X | X | |
| T359 | Cdk-7, HOG1-1 | X | X | NC | C | C | C |
| S372 | GIN4-5, PKA-1 | ? | ? | NC | C | C | X |
| S373 | GIN4-6 | NC | C | NC | C | C | ? |
| S396 | NC | NC | NC | ? | ? | C | |
| S413 | ? | X | X | C | NC | C | |
| S415 | C | C | C | NC | C | ? | |
| S559 | NC | X | NC | NC | C | X | |
| S561 | ? | C | NC | NC | C | C | |
| S583 | NC | X | NC | X | C | ? | |
| Coverage of predicted kinase sites not found by mass spectrometry | |||||||
| Asynchronous 1 | Arrested 1 | Heat Stress 1 | Asynchronous 2 | Arrested 2 | Heat Stress 2 | ||
| S024 | GIN4-1 | C | C | NC | C | C | C |
| T183 | GIN4-2 | NC | NC | NC | C | C | C |
| S271 | Cdk-5 | C | C | NC | C | C | C |
| S373 | GIN4-6 | C | C | NC | C | C | ? |
| T420 | Cdk-8 | NC | C | NC | C | C | C |
| S556 | GIN4-7 | NC | C | NC | NC | NC | C |
| S637 | GIN4-8, PKA-2 | NC | NC | NC | NC | NC | NC |
?, phosphorylated with low confidence, <50%. Blue X, phosphorylated with medium confidence, 50–90%. Green X, phosphorylated with high confidence, >90%. Red X, phosphorylated with high confidence with a lot more hits than in asynchronous. C, region covered by mass spectrometry. NC, region not covered by mass spectrometry.
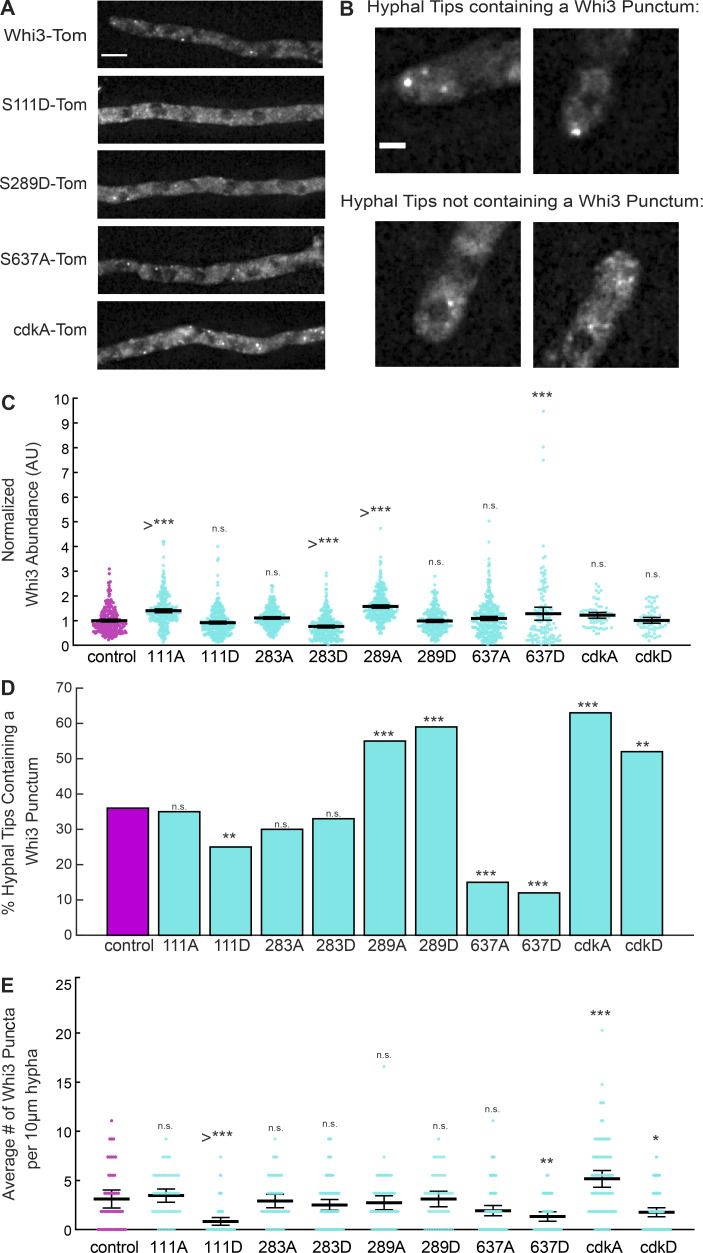
We found a spectrum of phenotypes that could be assigned into four distinct classes: complete loss of function with defects in both nuclear cycling and polarity, partial loss of function in nuclear cycling or polarity, or no phenotype (Table 2, Fig. 2, Fig. 3, and Fig. 4). Note, in order to highlight and contrast separation of function alleles and still allow comparison of all mutants in a given phenotype, the figures will sometimes be referenced out of a linear order. Importantly, all mutants are comparably expressed to wild-type protein, indicating that the defects are not due to misexpression of the mutant alleles (Fig. 4 C). The cell cycle defects impacted synchrony rather than progression through the division cycle per se (Fig. 2 D). In some cases, phenotypes were associated with the loss of detectable Whi3 condensates, but in other cases condensates appeared normal but were clearly not functional (Fig. 4). Similarly, in some but not all alleles, we found changes in mRNA levels that could explain mutant phenotypes. Below, we discuss the most interesting cases of clear separation of function alleles in detail and then examine the associations between localization and function.
Table 2. Summary of cellular phenotypes and Whi3 localization.
| Polarity | Tip Puncta | Asynchrony | Hyphal Puncta | |
|---|---|---|---|---|
| Control | + | + | + | + |
| 111A | + | + | + | + |
| 111D | + | - | - | -- |
| 283A | + | + | + | + |
| 283D | + | + | + | + |
| 289A | - | ++ | + | + |
| 289D | -- | ++ | - | + |
| 637A | -- | -- | + | + |
| 637D | -- | -- | - | -- |
| cdkA | - | ++ | - | ++ |
| cdkD | -- | ++ | - | - |
Figure 2.
Single point mutants at sites of Whi3 phosphorylation can increase nuclear synchrony. (A) Nuclei in wild-type A. gossypii cells go through the cell cycle asynchronously but can become more synchronous in certain genetic backgrounds. Nuclei cartooned in cell cycle progression shown with spindle pole bodies. (B) Representative images of SPB labeling for synchrony scoring in control (asynchronous), 111D (increased synchrony mutant), and 637A (asynchronous mutant). SPBs are in red and nuclei are in cyan. Hyphal outlines are dotted lines and representative nuclei of each nuclear cycle stage are indicated with arrows in control cell. Images are maximum intensity projections. Scale bar, 5 µm. (C) Synchrony indices for Whi3 mutants as compared with control cells. n > 180 for all conditions. Bars denote SE. *, P < 0.05. (D) Percent of nuclei in each phase of the nuclear cycle in control and Whi3 mutant strains. n > 180 for all conditions. G1 nuclei are orange bars, S/G2 nuclei are blue bars, and M nuclei are pink bars. Synchrony phenotype for each strain is represented by cartoon image below strain name. n.s., not significant; SPB, spindle pole body.
Figure 3.
Single point mutants at potential sites of Whi3 phosphorylation can disrupt cellular polarity and lateral branching. (A) Representative images to show gross morphology of control and Whi3 mutant strains grown on agar pads. Cartoon representation of polarity phenotype for each strain is shown below each strain. Scale bar, 50 µm. (B) Montage of cells growing on plate for control (normal polarity), 111D (normal polarity mutant), and 637A (disrupted polarity mutant). Interval between frames is 108 min. Scale bar, 50 µm. (C) Representative images of actin at hyphal tips in control and selected Whi3 mutant strains. Images are maximum intensity projections. Scale bar, 2 µm. (D) Length of polarized actin zone along hyphal long axis in control and Whi3 mutant strains. n > 31 for all conditions. *, P < 0.05 by one-way ANOVA with multiple comparisons. **, P < 0.005; ***, P < 0.0005. Bars, 95% confidence interval.
Figure 4.
Single point mutants at potential sites of Whi3 phosphorylation can strongly influence Whi3 localization and assembly. (A) Representative images of Whi3 protein in control or Whi3 mutant strains localized in the hypha. Images are maximum intensity projections. Scale bar, 5 µm. Tom, Tomato. (B) Representative images of hyphal tips containing or not containing Whi3-tomato puncta. Images are maximum intensity projections. Scale bar, 2 µm. (C) Normalized Whi3 abundance in control and Whi3 mutant cells under different growth conditions. n > 62 for all conditions. *, P < 0.05 by one-way ANOVA with multiple comparisons. Bars, 95% confidence interval. (D) Percent of hyphal tips in control and Whi3 mutant strains containing a Whi3 punctum. n > 167 for all conditions. *, P < 0.05 by N-1 χ2 test with Benjamini–Hochberg correction. (E) Average number of Whi3 puncta per 10-µm hyphal segment in control and Whi3 mutant strains. *, P < 0.05 by one-way ANOVA with multiple comparisons. **, P < 0.005; ***, P < 0.0005. Bars, 95% confidence interval.
Separation of function mutants specific for asynchrony or cell polarity
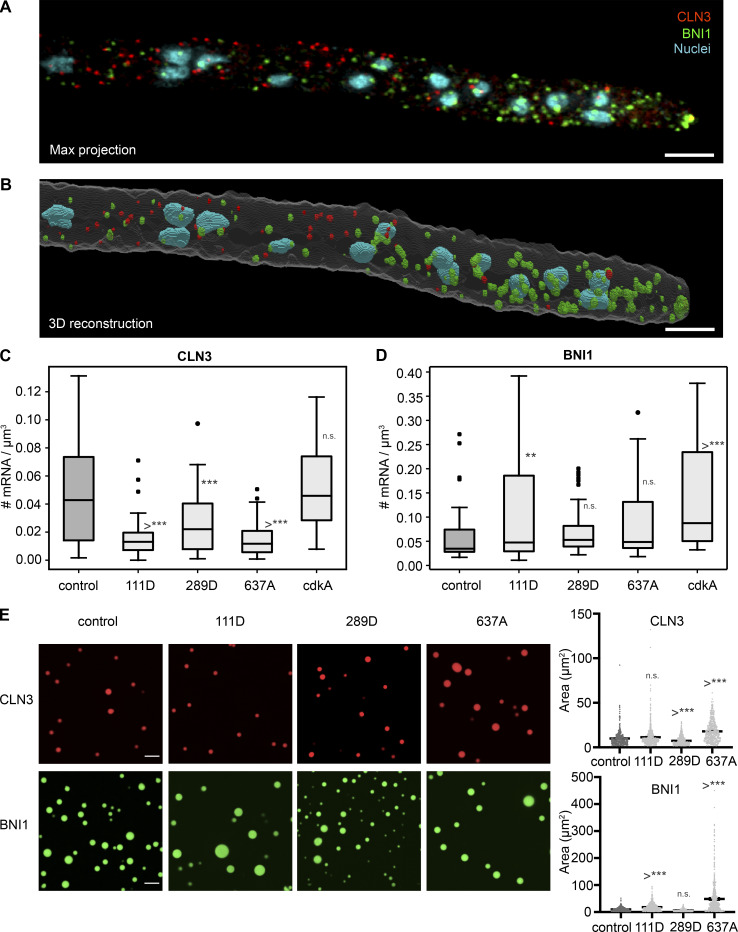
Two clear separation of function alleles emerged, supporting that phosphoregulation is a key cue for functional specialization of Whi3 condensates. One partial loss of function allele specifically impacted nuclear asynchrony; cells expressing S111D mutations are significantly more synchronous than wild-type cells (Fig. 2 C) but show normal branching patterns (Fig. 3, A and B). S111D is a predicted Cdk site that we detected as a phosphorylation event more frequently in both synchronized cells as well as in heat-stressed cells. Consistent with the loss of asynchrony phenotype, S111D mutants showed fewer Whi3 puncta associated with nuclei (Fig. 4 E) and a lower overall level of CLN3 mRNA, indicating a failure to properly regulate the stability of this transcript (Fig. S1 C). There is also a modest but statistically significant decrease in tip-associated puncta but presumably still enough assembly to support normal polarity patterning (Fig. 3 and Fig. 4 D), no difference in the length of the polarized actin zone in these cells compared with controls (Fig. 3 D), and a slightly elevated overall level of BNI1 mRNA (Fig. S1 D). Thus, a single residue change is sufficient to specifically impact Whi3 in the cell cycle without impacting functions in cell polarity.
Figure S1.
Overall cytoplasmic concentration of BNI1 and CLN3. (A) Max projection of representative smFISH image, contrasted for visibility. BNI1 was labeled with CY5, CLN3 was labeled with TAMRA, and nuclei were DAPI-stained. Scale bar, 5 µm. (B) 3D reconstruction generated in ImageTank of the cell in A. Masks of the hypha (white), BNI1 spots (green), CLN3 spots (red), and nuclei (blue) are visible. Scale bar, 5 µm. (C) Overall cytoplasmic concentration of BNI1. Each datapoint is the overall concentration of a single hypha. For each strain, n > 33 hyphae. *, P < 0.05 by one-way ANOVA with multiple comparisons. Bars denote 95% confidence interval. (D) Box plot of overall cytoplasmic concentration of CLN3. Each datapoint is the overall concentration of a single hypha. Scale bar, 10 µm. For each strain, n > 33 hyphae. *, P < 0.05 by one-way ANOVA with multiple comparisons. (E) Representative images of control and mutant Whi3 protein phase separated with CLN3 and BNI1 RNA in vitro. RNA channel is labeled. Area of CLN3 or BNI1 droplets phase separated with different Whi3 protein in vitro. *, P < 0.05 by one-way ANOVA with multiple comparisons. n > 500 for all conditions. Bars denote mean and 95% confidence interval. **, P < 0.005; ***, P < 0.0005.
The second separation of function allele we identified was S637A. S637 is located within the RNA-recognition motif (Fig. 1 F), of Whi3 and is a conserved, putative PKA consensus site (Mizunuma et al., 2013). The S637A mutant has normal nuclear asynchrony (Fig. 2 C) but grossly disrupted polarity (Fig. 3, A and B). There is a 50% reduction in tips with Whi3 puncta compared with wild-type cells in the S637A allele, consistent with the substantial polarity problems (Fig. 3 and Fig. 4) and an increase in length of the polarized actin zone at tips (Fig. 3 D). Overall BNI1 mRNA levels are not disturbed in this mutant, although the levels are more variable cell to cell (Fig. S1 D). This indicates that the observed polarity initiation problems are unlikely a consequence of destabilizing BNI1. Furthermore, there is no reduction in Whi3 puncta in the vicinity of nuclei (Fig. 4 E), consistent with Whi3 being able to support its normal role in regulating cell cycle progression. Curiously, in these cells there is a decrease in overall CLN3 mRNA abundance, indicating that lowering CLN3 levels is not sufficient to generate synchrony (Fig. S1 C). The S637A mutants have specifically lost the ability to regulate polarity without impacting nuclear cycling.
Thus, separation of function alleles was identified for both the key processes controlled by Whi3 condensates in A. gossypii. Remarkably, these single point mutations were each able to recapitulate what was seen in the full polyQ deletion mutant for each process with respect to cellular phenotype and Whi3 localization (Lee et al., 2013, 2015). But unlike the polyQ mutants, only a specific subset of Whi3 assemblies was lost, and the corresponding cellular function was impaired. This indicates that it is possible to genetically separate and independently regulate these distinct functions of Whi3.
There has been an emerging understanding of how post-translational modification can be used as a cue to regulate assembly or disassemble of condensate structures in other systems (Banani et al., 2016, Hofweber and Dormann, 2019). To our knowledge, however, it has not been shown that post-translational modifications can be used to specify different functions for condensates that share common components. Our results indicate that beyond the emergent properties of weak, multivalent interactions, there are likely to be specific and switchable interactions that are important for assembly. Post-translational modifications may lead to different client proteins that impart functional specificity to condensates.
Putative Cdk-target residues are important for Whi3 function
Initially, we were concerned that single residues may be insufficient to generate a phenotype, so in parallel, we made mutants where eight predicted Cdk sites (T34, S39, S111, S249, S271, T314, T359, and T420) were mutated to be phosphomimetic (Ser/Thr to Asp) or nonphosphorylatable (to Ala). We termed these mutants cdkA and cdkD. Of these eight sites, five were found to be phosphorylated in one or more conditions in our mass spectrometry (T34, S39, S111, S249, and T359), while the other three sites were not detected in our conditions. Both the cdkA and the cdkD mutant showed extreme defects in both polarity and nuclear asynchrony (Fig. 2 and Fig. 3). Surprisingly, both the strains showed an increase in the amount of Whi3 puncta at hyphal tips despite significant defects in generating lateral branches. This supports a role for Cdk/cyclin in cell polarity, which would be predicted based on the localization of Cln1/2 cyclin protein at tips (Gladfelter et al., 2006). Thus, presence of a puncta at a tip is not sufficient for normal polarity and suggests that condensates need to be properly regulated by Cdk to function.
Interestingly, though both strains showed pronounced defects in nuclear synchrony, the cdkA strains had increased Whi3 hyphal puncta while the cdkD strains had very few. These changes in the frequency of interregion puncta suggest that phosphoregulation is an important cue for the assembly and dissolution of Whi3 puncta. Notably, similar loss of function phenotypes can arise from seemingly opposing mutations when condensate assembly/disassembly are perturbed. When we examined overall CLN3 and BNI1 levels in the cdkA mutant we found that levels of CLN3 mRNA were unchanged and BNI1 levels were increased (Fig. S1, C and D), suggesting that these Whi3 proteins are likely able to bind mRNA. These data indicate that simply the presence of the Whi3 protein and target mRNA is insufficient for normal activity in controlling the division cycle.
Whi3 residues phosphorylated during heat stress do not affect polarity or nuclear cycle regulation
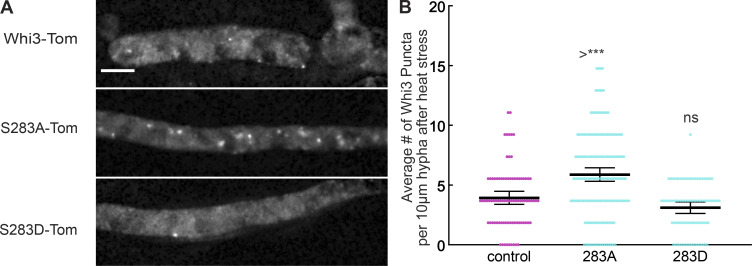
One site, S283, was specifically targeted during heat stress. Mutations at site S283 did not have any effect on nuclear synchrony or polarized growth (Figs. 2 and 3). Similarly, there was no difference in the number of hyphal tips containing a Whi3 punctum between wild type and either of these mutants (Fig. 4 D), consistent with the fact that there was no polarity defect in these strains. With respect to hyphal Whi3 puncta, S283D was indistinguishable from wild type, consistent with its having no problem maintaining nuclear asynchrony (Fig. 4 E). S283A, however, had slightly fewer hyphal puncta than control in spite of having no defect in nuclear synchrony (Fig. 4 E).
We hypothesize that a subset of hyphal Whi3 puncta may be used to regulate other Whi3 phenotypes related to environmental stress response, and that these puncta are absent in the S283 strain without affecting Whi3-regulated nuclear asynchrony. When these cells were imaged under heat stress conditions, we saw more Whi3 puncta in the S283A cells compared with control (Fig. S2), consistent with this residue playing a role in regulating Whi3 assemblies involved in a heat stress response. In S283D cells, there was no change in the number of Whi3 puncta compared with control following heat stress (Fig. S2). Taken together, these results raise the possibility that dephosphorylation at S283 during environmental stress might be used to promote a stress-specific Whi3 assembly or even to promote Whi3 recruitment to stress granules.
Figure S2.
Residue S283 regulates Whi3 response to heat stress. (A) Representative images of Whi3 protein in control, 283A, and 283D strains localized in the hypha following heat stress. Images are maximum intensity projections. Scale bar, 5 µm. (B) Average number of Whi3 puncta per 10 µm hyphal segment in control, 283A, and 283D strains localized in the hypha following heat stress. n > 75 for all conditions. *, P < 0.05 by one-way ANOVA with multiple comparisons. ***, P < 0.0005. Bars denote 95% confidence interval.
Alleles with loss of function in polarity and nuclear asynchrony
In addition to the CDK mutants, there were two other alleles that showed complete loss of function in asynchrony and cell polarity: S289D and S637D. Interestingly, while the S637A strain does show separation of Whi3 function, the corresponding phosphomimetic S637D strain has both aberrant polarity and a more synchronous nuclear cycle, which cannot be easily explained by the site being used exclusively to regulate Whi3 for its function in polarized growth. The S637D cells have also lost both tip and hyphal puncta. There are a number of potential interpretations of this, but one possibility is that phosphorylation of S637 is required for polarized growth but not for nuclear asynchrony. However, the S637D mutation is not an appropriate mimic for a true phosphorylation event and has created a misfunctional protein that can no longer fulfill any of its normal functions.
Cells expressing the phosphomimetic S289D variant of Whi3 are highly synchronous (Fig. 2 C) and show substantial polarity defects with a longer zone of polarized actin (Fig. 3, A and D). Notably, these phenotypes are not associated with Whi3’s inability to form puncta as in fact there are still hyphal puncta and even more tips with puncta in this mutant (Fig. 4, D and E). S289D cells show a modest decrease in overall CLN3 concentration and no change in overall BNI1 levels (Fig. S1, C and D), consistent with the idea that in these cells, Whi3 is forming complexes with its target mRNA that are nonetheless not fully functional. We hypothesize that regulated phosphorylation at S289 is important for recruitment of other factors that are needed to make Whi3 complexes fully functional for cell polarity and nuclear asynchrony.
Loss of function not always associated with loss of condensation
Previous work has shown that when the polyQ tract is removed from Whi3 (ΔpolyQ), the protein is found far less often in puncta (both hyphal and at tips), because its ability to form assemblies has been impaired, and in vitro this is seen as an inability to undergo a liquid–liquid phase separation with target RNAs (Lee et al., 2013, 2015; Zhang et al., 2015). The situation is far more complex with the phosphomutants, where a substantial fraction of the mutants still forms higher-order assemblies in cells and in some cases is even more abundant than the condensates made by wild-type protein. As noted above, both the S289 and Cdk alleles all form abundant tip puncta but cannot branch normally, and the S289D and cdkA mutants show normal to abundant hyphal puncta but a nuclear synchrony phenotype. This raises the possibility that the purpose of forming these structures is not simply to bind or localize RNA but likely involves additional roles such as regulation of RNA expression. This is consistent with the fact that Whi3 mutant alleles unable to support in vivo function are able to phase separate with target mRNA in vitro (Fig. S1 E). These alleles demonstrate an important role for phosphoregulation beyond controlling the assembly state but also in the function potentially thought mediating specific protein or RNA interactions.
Conclusions
In conclusion, this study shows that phosphorylation of a single residue can specifically regulate a subset of condensate assemblies built from a common scaffold protein. Furthermore, in addition to stabilizing or destabilizing the assemblies themselves, phosphorylation can also be used to regulate the activity of assemblies, possibly through the recruitment of other client proteins. Overall, this work advanced our understanding of the molecular grammar used to regulate condensates in the cell.
Materials and methods
Strain construction
To create Whi3 mutant strains tagged with tomato, geneblocks containing the mutations of interest were ordered from IDT (Table 3). These geneblocks were then used to replace the relevant portions of Whi3 in AGB993 using Gibson cloning to amplify the geneblocks and relevant portions of the plasmid (Table 4). Constructs were sequenced across the Whi3 region of the plasmid to ensure that no additional mutations had accrued during the cloning process (Table 5). Each plasmid was then cut by SacII, XhoI, and NdeI. The 6.377-kb fragments containing the mutant Whi3, the fluorescent tag, and the Nourseothricin sulfate resistance marker were separated and purified using an agarose gel and integrated into A. gossypii using protocols described previously (Wendland and Walther, 2005; Table 6).
Table 3. Geneblocks.
| Geneblocks | |
|---|---|
| S111A | GATGGGGGCGGAATCGCAAGGGGGCGGCGCAAGCAATAACAACAACAACCCCGCGAACCCGGGCAGCACGGCCAACAATAATAGTAATAACGTCAACATGAACAGCATCGGTGGCGGGGCGAGCTTGGGTGCCGGTAGTGGGGCCACCGGCAGTATTTCGGGAACGAAGGGCATGAACAACAGCCATGCGCCGCTCCATATTGCCACGATGCTGAACACCTTGTCGATGAACTCGAATCCGCCATCCCAACAGCAGTCCAATGTACAGGGCCCTTACTTGGTGCGGTTGCAGAATGTTCCGAAGGACACTACCTTGCGGGAATGCCATGCTTTGTTTGCATTGGCGCACGGCGTGTTATCTATCGAGTTGT |
| S111D | GATGGGGGCGGAATCGCAAGGGGGCGGCGCAAGCAATAACAACAACAACCCCGCGAACCCGGGCAGCACGGCCAACAATAATAGTAATAACGTCAACATGAACAGCATCGGTGGCGGGGCGAGCTTGGGTGCCGGTAGTGGGGCCACCGGCAGTATTTCGGGAACGAAGGGCATGAACAACAGCCATGACCCGCTCCATATTGCCACGATGCTGAACACCTTGTCGATGAACTCGAATCCGCCATCCCAACAGCAGTCCAATGTACAGGGCCCTTACTTGGTGCGGTTGCAGAATGTTCCGAAGGACACTACCTTGCGGGAATGCCATGCTTTGTTTGCATTGGCGCACGGCGTGTTATCTATCGAGTTGT |
| S283A | TACGTTGAAGTGGTTGACGAGCTCACGCAACAGCAGATACCGTTCCAGACTCAAATGCAAATGCACCAGGGCTCCCCACCAGCCCCCACCCATGTCACTGCATATCAACAGCCTTTGCTATCTGCTTCCGGCGTGGTTAGCCCACCGCAATCAGCTTCTAGTGTGAAGAGACCCGCGCTTTTGGTCCAACGTTCCAGGTTCTCATTCACGGATCCGTTTTCTAGCGAGCAAACTAATATGGGCTCACAGCAGCCAGATCTAATTACGACGCCGTTGAAAGGTCACCAGGACACTGGGAAGTCGTTTTTATTGATGGAAAGTGATGAGATTAACGATAGTATATGGGGTAATGGAACCGGCATTCC |
| S283D | TACGTTGAAGTGGTTGACGAGCTCACGCAACAGCAGATACCGTTCCAGACTCAAATGCAAATGCACCAGGGCTCCCCACCAGCCCCCACCCATGTCACTGCATATCAACAGCCTTTGCTATCTGCTTCCGGCGTGGTTAGCCCACCGCAATCAGCTTCTAGTGTGAAGAGACCCGACCTTTTGGTCCAACGTTCCAGGTTCTCATTCACGGATCCGTTTTCTAGCGAGCAAACTAATATGGGCTCACAGCAGCCAGATCTAATTACGACGCCGTTGAAAGGTCACCAGGACACTGGGAAGTCGTTTTTATTGATGGAAAGTGATGAGATTAACGATAGTATATGGGGTAATGGAACCGGCATTCC |
| S289A | AACAGCAGATACCGTTCCAGACTCAAATGCAAATGCACCAGGGCTCCCCACCAGCCCCCACCCATGTCACTGCATATCAACAGCCTTTGCTATCTGCTTCCGGCGTGGTTAGCCCACCGCAATCAGCTTCTAGTGTGAAGAGACCCAGCCTTTTGGTCCAACGTGCGAGGTTCTCATTCACGGATCCGTTTTCTAGCGAGCAAACTAATATGGGCTCACAGCAGCCAGATCTAATTACGACGCCGTTGAAAGGTCACCAGGACACTGGGAAGTCGTTTTTATTGATGGAAAGTGATGAGATTAACGATAGTATATGGGGTAATGGAACCGGCATTCCTTCCAGCATAAGTGGTTTG |
| S289D | AACAGCAGATACCGTTCCAGACTCAAATGCAAATGCACCAGGGCTCCCCACCAGCCCCCACCCATGTCACTGCATATCAACAGCCTTTGCTATCTGCTTCCGGCGTGGTTAGCCCACCGCAATCAGCTTCTAGTGTGAAGAGACCCAGCCTTTTGGTCCAACGTGACAGGTTCTCATTCACGGATCCGTTTTCTAGCGAGCAAACTAATATGGGCTCACAGCAGCCAGATCTAATTACGACGCCGTTGAAAGGTCACCAGGACACTGGGAAGTCGTTTTTATTGATGGAAAGTGATGAGATTAACGATAGTATATGGGGTAATGGAACCGGCATTCCTTCCAGCATAAGTGGTTTG |
| S637A | AAATGCTGCTGCAGCTTTGCAAAATACTAATGGTATTTCACAAGTTGATTTATCTTTGTTGGCTAAAGTTCCTCCTCCCGCAAATCCAGCTGATCAGAACCCCCCTTGTAACACTCTCTATGTTGGTAACTTGCCGCCAGATGCAACTGAACAAGAATTGCGCCAGTTATTCTCTAGTCAGAAGGGATTCAGGAGATTGGCCTTTAGGAATAAAAATAACAACGGTAATGGCCATGGTCCAATGTGCTTTGTTGAGTTCGAGGACGTTGCGCATGCAACGAGGGCATTGGCTGAATTATACGGTAGTCAATTGGCGCGTACAAGCGGCACCCACAATAATAAAGGTGGAATTAGGTTGAGTTTCTCTAAGAACCCATTGGGTGTCAGGGGTCCCAACAGCAGAAGAG |
| S637D | AAATGCTGCTGCAGCTTTGCAAAATACTAATGGTATTTCACAAGTTGATTTATCTTTGTTGGCTAAAGTTCCTCCTCCCGCAAATCCAGCTGATCAGAACCCCCCTTGTAACACTCTCTATGTTGGTAACTTGCCGCCAGATGCAACTGAACAAGAATTGCGCCAGTTATTCTCTAGTCAGAAGGGATTCAGGAGATTGGACTTTAGGAATAAAAATAACAACGGTAATGGCCATGGTCCAATGTGCTTTGTTGAGTTCGAGGACGTTGCGCATGCAACGAGGGCATTGGCTGAATTATACGGTAGTCAATTGGCGCGTACAAGCGGCACCCACAATAATAAAGGTGGAATTAGGTTGAGTTTCTCTAAGAACCCATTGGGTGTCAGGGGTCCCAACAGCAGAAGAG |
| cdkA | CAAGCCTGACTCAAGTGGTTCGCAGGACCGCCTGGTATTCCGCAGACGCGAGGGATCGGGAGGATCGAGAGCGATGTCGCTGGTTAACAGTCACTCGTCTGCGAGCGTGGAAAACGCTGCGTACAACCTACACCGCGCATTCAGCTCTAGCACGGAGAACGTGGGGCACATGGCGCCCAGCAACTCGGCGCCGTTGCACCACTCCACGGTGGTAGCGATGGGGGCGGAATCGCAAGGGGGCGGCGCAAGCAATAACAACAACAACCCCGCGAACCCGGGCAGCACGGCCAACAATAATAGTAATAACGTCAACATGAACAGCATCGGTGGCGGGGCGAGCTTGGGTGCCGGTAGTGGGGCCACCGGCAGTATTTCGGGAACGAAGGGCATGAACAACAGCCATGCGCCGCTCCATATTGCCACGATGCTGAACACCTTGTCGATGAACTCGAATCCGCCATCCCAACAGCAGTCCAATGTACAGGGCCCTTACTTGGTGCGGTTGCAGAATGTTCCGAAGGACACTACCTTGCGGGAATGCCATGCTTTGTTTGCATTGGCGCACGGCGTGTTATCTATCGAGTTGTCGAGCTTCCAGCAGTACGCCGAGCGATCTCAGACGTCTGGCCAGGAGTCCACAAATTACATCGTTGCCAAATTCGATTCTTTGCATTTGGCGTGCCAGTATGCGACTATTCTTGATGAGAAGGCTCAGATATTCGGGCCCAGCTTTCCGTTCAAAACCTACGTTGAAGTGGTTGACGAGCTCACGCAACAGCAGATACCGTTCCAGACTCAAATGCAAATGCACCAGGGCGCGCCACCAGCCCCCACCCATGTCACTGCATATCAACAGCCTTTGCTATCTGCTTCCGGCGTGGTTGCGCCACCGCAATCAGCTTCTAGTGTGAAGAGACCCAGCCTTTTGGTCCAACGTTCCAGGTTCTCATTCACGGATCCGTTTTCTAGCGAGCAAACTAATATGGGCTCACAGCAGCCAGATCTAATTACGGCGCCGTTGAAAGGTCACCAGGACACTGGGAAGTCGTTTTTATTGATGGAAAGTGATGAGATTAACGATAGTATATGGGGTAATGGAACCGGCATTCCTTCCAGCATAAGTGGTTTGACGACTTCGCAGCCACCAGCGCCTCACTTGGAGTGGGGTACCACGGGAAGGCGCCAAAGTTCAACTTTTTATCCTTCGCAGTCGAACACAGAAATACCACCTATGCATTTAACGGGTCAAGTGCAATCTTCACAACTAGCAACTGGTTTGCAACAACCGCTACCACAGCCGCAACGTCAGTCTTTGTCATATAACTTGGTAGCGCCGCTATCCTCCGATATGAACTTGCCACCTCAGTCTTCACAGGGCGGCATATTACCGCATCAGGCGCCAGCGC |
| cdkD | CAAGCCTGACTCAAGTGGTTCGCAGGACCGCCTGGTATTCCGCAGACGCGAGGGATCGGGAGGATCGAGAGCGATGTCGCTGGTTAACAGTCACTCGTCTGCGAGCGTGGAAAACGCTGCGTACAACCTACACCGCGCATTCAGCTCTAGCACGGAGAACGTGGGGCACATGGACCCCAGCAACTCGGACCCGTTGCACCACTCCACGGTGGTAGCGATGGGGGCGGAATCGCAAGGGGGCGGCGCAAGCAATAACAACAACAACCCCGCGAACCCGGGCAGCACGGCCAACAATAATAGTAATAACGTCAACATGAACAGCATCGGTGGCGGGGCGAGCTTGGGTGCCGGTAGTGGGGCCACCGGCAGTATTTCGGGAACGAAGGGCATGAACAACAGCCATGACCCGCTCCATATTGCCACGATGCTGAACACCTTGTCGATGAACTCGAATCCGCCATCCCAACAGCAGTCCAATGTACAGGGCCCTTACTTGGTGCGGTTGCAGAATGTTCCGAAGGACACTACCTTGCGGGAATGCCATGCTTTGTTTGCATTGGCGCACGGCGTGTTATCTATCGAGTTGTCGAGCTTCCAGCAGTACGCCGAGCGATCTCAGACGTCTGGCCAGGAGTCCACAAATTACATCGTTGCCAAATTCGATTCTTTGCATTTGGCGTGCCAGTATGCGACTATTCTTGATGAGAAGGCTCAGATATTCGGGCCCAGCTTTCCGTTCAAAACCTACGTTGAAGTGGTTGACGAGCTCACGCAACAGCAGATACCGTTCCAGACTCAAATGCAAATGCACCAGGGCGACCCACCAGCCCCCACCCATGTCACTGCATATCAACAGCCTTTGCTATCTGCTTCCGGCGTGGTTGACCCACCGCAATCAGCTTCTAGTGTGAAGAGACCCAGCCTTTTGGTCCAACGTTCCAGGTTCTCATTCACGGATCCGTTTTCTAGCGAGCAAACTAATATGGGCTCACAGCAGCCAGATCTAATTACGGACCCGTTGAAAGGTCACCAGGACACTGGGAAGTCGTTTTTATTGATGGAAAGTGATGAGATTAACGATAGTATATGGGGTAATGGAACCGGCATTCCTTCCAGCATAAGTGGTTTGACGACTTCGCAGCCACCAGACCCTCACTTGGAGTGGGGTACCACGGGAAGGCGCCAAAGTTCAACTTTTTATCCTTCGCAGTCGAACACAGAAATACCACCTATGCATTTAACGGGTCAAGTGCAATCTTCACAACTAGCAACTGGTTTGCAACAACCGCTACCACAGCCGCAACGTCAGTCTTTGTCATATAACTTGGTAGACCCGCTATCCTCCGATATGAACTTGCCACCTCAGTCTTCACAGGGCGGCATATTACCGCATCAGGCGCCAGCGC |
Table 4. Primers to create Gibson fragments.
| Name | Description | Sequence |
|---|---|---|
| AGO1963 | S637X_5' F | ccctttcgtcTCGCGCGTTTCGGTGATG |
| AGO1964 | S637X_5' R | gcaaagctgcAGCAGCATTTTTAGGGCCGTTG |
| AGO1965 | S637X_3' F | gggtcccaacAGCAGAAGAGGTGGTGCTACC |
| AGO1966 | S637X_3' R | aaacgcgcgaGACGAAAGGGCCTCGTGATAC |
| AGO1967 | S289X_5' F | ccctttcgtcTCGCGCGTTTCGGTGATG |
| AGO1968 | S289X_5' R | ctggaacggtATCTGCTGTTGCGTGAGCTCG |
| AGO1969 | S289X_3' F | cttccagcatAAGTGGTTTGACGACTTCGC |
| AGO1970 | S289X_3' R | aaacgcgcgaGACGAAAGGGCCTCGTGATAC |
| AGO1971 | S283X_5' F | ccctttcgtcTCGCGCGTTTCGGTGATG |
| AGO1972 | S283X_5' R | tcgtcaaccaCTTCAACGTAGGTTTTGAACGGAAAG |
| AGO1973 | S283X_3' F | ggtaatggaaCCGGCATTCCTTCCAGCATAAG |
| AGO1974 | S283X_3' R | aaacgcgcgaGACGAAAGGGCCTCGTGATAC |
| AGO1975 | S111X_5' F | ccctttcgtcTCGCGCGTTTCGGTGATG |
| AGO1976 | S111X_5' R | cttgcgattcCGCCCCCATCGCTACCAC |
| AGO1977 | S111X_3' F | cgtgttatctATCGAGTTGTCGAGCTTCC |
| AGO1978 | S111X_3' R | aaacgcgcgaGACGAAAGGGCCTCGTGATAC |
| AGO1979 | cdkmutant_5' F | ccctttcgtcTCGCGCGTTTCGGTGATG |
| AGO1980 | cdkmutant_5' R | aaccacttgaGTCAGGCTTGCGTGCGTTC |
| AGO1981 | cdkmutant_3' F | accgcatcagGCGCCAGCGCAGACGCAG |
| AGO1982 | cdkmutant_3' R | aaacgcgcgaGACGAAAGGGCCTCGTGATACGC |
Table 5. Plasmids.
| Plasmids | |
|---|---|
| AGB1020 | pRS416 Whi3S637A-tomato-NAT |
| AGB1021 | pRS416 Whi3S637D-tomato-NAT |
| AGB1022 | pRS416 Whi3S289A-tomato-NAT |
| AGB1023 | pRS416 Whi3S289D-tomato-NAT |
| AGB1024 | pRS416 Whi3S283A-tomato-NAT |
| AGB1025 | pRS416 Whi3S283D-tomato-NAT |
| AGB1026 | pRS416 Whi3S111A-tomato-NAT |
| AGB1027 | pRS416 Whi3S111D-tomato-NAT |
| AGB1028 | pRS416 Whi3cdkmutantA-tomato-NAT |
| AGB1029 | pRS416 Whi3cdkmutantD-tomato-NAT |
Table 6. Strains used in this study.
| Strains | |
|---|---|
| 834 | ΔlΔt-Whi3-tomato::NAT |
| 853 | ΔlΔt-Whi3(S637A)-tomato::NAT |
| 854 | ΔlΔt-Whi3(S637D)-tomato::NAT |
| 855 | ΔlΔt-Whi3(S289A)-tomato::NAT |
| 856 | ΔlΔt-Whi3(S289D)-tomato::NAT |
| 857 | ΔlΔt-Whi3(S283A)-tomato::NAT |
| 858 | ΔlΔt-Whi3(S283D)-tomato::NAT |
| 859 | ΔlΔt-Whi3(S111A)-tomato::NAT |
| 860 | ΔlΔt-Whi3(S111D)-tomato::NAT |
| 861 | ΔlΔt-Whi3(cdkmutA)-tomato::NAT |
| 862 | ΔlΔt-Whi3(cdkmutD)-tomato::NAT |
Whi3 protein imaging and analysis
A. gossypii cells were grown for 12.5 h shaking at 150 rpm in Ashbya full media (AFM) at 30°C with appropriate selection, collected by centrifugation for 2 min at 500 rpm, and resuspended in 2× low fluorescence minimal media. Heat-stressed cells were shifted to 42°C at 10.5 h. Cells were arrested with 7.5 µg/ml nocodazole at 10.5 h. Cells were mounted on 1.4% agarose gel pads made with low fluorescence minimal media, sealed with Valap, and then imaged using a widefield microscope (Nikon Eclipse TI stage) with a Plan Apo λ 60×/1.40 oil Ph3 DM objective and an Andor Zyla 4.2 plus VSC-06258 camera. Images were deconvolved using 12 iterations of the Lucy–Richardson algorithm in Nikon Elements software and then processed for display using Fiji.
To measure Whi3 abundance, hyphal segments were traced in the phase channel in a middle plane to create regions of interest (ROIs). Z-series were used to create a sum intensity projection of the fluorescent channel, and then the mean fluorescent signal in each ROI was measured. These values were then averaged to generate Whi3 abundance in each strain. Abundances of mutant strains were then normalized to the control abundance. To measure hyphal tips containing Whi3 puncta, hyphal tips were marked in phase channel and then manually scored for the presence or absence of a Whi3 punctum in fluorescent channel. Mutant strains were compared with control using an N-1 χ2 test with Benjamini–Hochberg correction. To measure the average number of Whi3 puncta in hyphae, 10-µm hyphal segments were traced in phase channel, and then the number of Whi3 puncta in each ROI was counted manually. Mutant strains were compared with control using ANOVA with multiple comparison correction.
Mass spectrometry
For mass spectrometry, A. gossypii cells were grown for 16.5 h shaking in AFM at 30°C with appropriate selection. Heat-stressed cells were shifted to 42°C at 14.5 h. Cells were arrested with 7.5 µg/ml nocodazole at 14.5 h. Cells were collected via vacuum filtration and rinsed with PBS before being resuspended in 2× NP-40 buffer (6 mM Na2HPO4, 4 mM NaH2PO4 × H2O, 1% NP-40, 150 mM NaCl, 2 mM EDTA, 50 mM NaF, 4 µg/ml leupeptin, 0.1 mM Na3VO4 + fresh 1× EDTA-free protease inhibitor, 1 mM PMSF, 0.01 mg/ml leupeptin, and 50 mM β-glycerophosphate). Resuspended cells were flash-frozen in liquid nitrogen, then lysed using a coffee grinder at 4°C. Lysate was clarified by spinning in a tabletop centrifuge at 13.K rpm and 4°C for 20 min. Supernatant was combined with IgG Sepharose beads and allowed to bind rotating at 4°C for 2 h. Beads were washed with IPP 150 buffer (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 0.1% NP-40) and resuspended in denaturing buffer (2% SDS, 50 mM Tris, pH 8.1, 50 mM NaCl, and 20% glycerol + fresh 2 mM DTT). The sample was incubated at 65°C for 20 min, then cooled to room temperature before adding 6 mM iodoacetamide and vortexed. The sample was incubated at room temperature in the dark for 1 h. DTT was then added to a final concentration of 5 mM to quench. Samples were run in SDS-PAGE, and bands corresponding to Whi3 by molecular weight were excised and differentially digested with trypsin, chymotrypsin, or proteinase-K, extracted, desalted by STAGE-tip extraction, and analyzed by microcapillary liquid chromatography-mass spectrometry/mass spectrometry using a Q-Exactive Plus mass spectrometer system as previously described (Kettenbach et al., 2018). The resulting mass spectra were data-searched using the Comet algorithm against the A. gossypii proteome with static modification of 57.02146 on Cys and dynamic modifications of 15.99491 on Met and 79.96633 on Ser, Thr, and Tyr residues using tryptic, chymotryptic, or no enzyme specificity and a mass tolerance of 1 D and filtered to a <1% false discovery rate using the target-decoy strategy by filtering post hoc (±3 ppm mass measurement accuracy and appropriate XCorr and deltaCorr values to achieve false discovery rate).
Two independent experiments were sent for mass spectrometry for each condition. Results for coverage of Whi3 in the first round were as follows: asynchronous: 51.3%, nocodazole arrested: 52.1%, heat stress: 7.1%. Results of coverage of Whi3 in the second round were as follows: asynchronous: 70.9%, nocodazole arrested: 73.9%, heat stress: 75.7%. In addition to the mapped sites, the protein sequence was manually scanned for predicted kinase consensus sites (Mok et al., 2010; Mizunuma et al., 2013) for kinases of interest.
Synchrony imaging and index
A. gossypii cells were grown for 16 h shaking in AFM at 30°C with appropriate selection, then fixed with 3.7% formaldehyde shaking at 30°C for 1 h. Cells were washed with PBS and resuspended in Solution A (100 mM K2PHO4, pH 7.5, and 1.2 M sorbitol) and digested with zymolyase at 37°C to remove cell walls. Cells were washed with Solution A and then spotted onto polylysine-treated wells on a slide. Cells were washed with PBS, blocked with BSA, and then incubated overnight with rat α-tubulin antibody at 4°C. The following day, cells were washed and incubated with fluorescently labeled secondary antibody + Hoechst. Cells were washed a final time and then covered with Prolong gold mounting medium for imaging. Cells were imaged using a widefield microscope (Nikon Eclipse TI stage) with a Plan Apo λ 100×/1.45 oil Ph3 DM objective and an Andor Zyla VSC-06258 camera. Images were deconvolved using 25 iterations of the Lucy–Richardson algorithm in Nikon Elements software and then processed using Fiji. Cell synchrony index was performed as described previously (Nair et al., 2010). Briefly, it is a method developed by our laboratory in collaboration with a statistician that allows us to compare the likelihood that adjacent nuclei are in the same cell cycle state compared with what would be expected by chance.
Actin zone length imaging and analysis
A. gossypii cells were grown for 15 h shaking in AFM at 30°C with appropriate selection and then fixed with 3.7% formaldehyde shaking at 30°C for 1 h. Cells were collected by centrifugation, washed with PBS, then incubated in PBS with Alexa Fluor Phalloidin 488 at 6.6 µM for 1 h. Cells were washed with PBS and mounted on glass slides with Prolong Gold mounting medium. Cells were imaged on a Zeiss LSM 880 AIRYSCAN with a Plan-Apochromat 63×/1.4 oil DIC M27 objective. Images were AIRYSCAN-processed using Zen software. Actin zone lengths were measured using Fiji. Mutant strains were compared with control using ANOVA with multiple comparison correction.
Cell branching imaging
A. gossypii spores were spotted onto agar plates made with AFM and appropriate selection. Cells were grown at 30°C and imaged on a Nikon Eclipse TS100 with a 10×/0.25 Ph1 ADL objective using an iDu Optics LabCam Microscope Adaptor and an iPhone 6 in LapseIt software.
Single molecule FISH (smFISH) imaging
RNA smFISH labeling of A. gossypii was performed as previously described (Lee et al., 2013) with minor adjustments. Briefly, A. gossypii cells were grown for 16 h shaking in AFM at 30°C with appropriate selection, then fixed with 3.7% formaldehyde shaking at 30°C for 1 h. Cells were collected by centrifugation, washed with cold Solution A, and resuspended in spheroplasting buffer (100 mM K2PHO4, pH 7.5, 1.2 M sorbitol, and 2 mM Vanadyl ribonucleoside complex), and digested with zymolyase at 37°C to remove cell walls.
All subsequent steps were performed in RNase-free conditions. Digested cells were washed with cold Solution A and then incubated in 70% EtOH at 4°C overnight. Cells were washed with SSC wash buffer, then resuspended in hybridization buffer (100 mg/ml dextran sulfate, 1 µg/ml Escherichia coli tRNA, 2mM Vanadyl ribonucleoside complex, 0.2 mg/ml BSA, 2× SSC, and 10% vol/vol deionized formamide) and incubated overnight at 37°C with 5-carboxytetramethylrhodamine (TAMRA) or cy5 conjugated RNA FISH probes (Stellaris LGC Biosearch Technologies) complementary to RNA of interest. Cells were washed with SSC wash buffer and incubated with Hoechst at room temperature for 30 min. Cells were washed a final time with SSC wash buffer and then mounted on glass slides with Prolong Gold mounting medium.
Cells were imaged using a widefield microscope (Nikon Eclipse TI stage) with a Plan Apo λ 100×/1.45 oil Ph3 DM objective and an Andor Zyla VSC-06258 camera.
Analysis of smFISH images
Images were deconvolved using 29 iterations of the Lucy–Richardson algorithm in Nikon Elements.
The modular image processing software ImageTank was used to analyze smFISH images and calculate RNA concentrations. The dataset of 100 multi-channel z-stacks was read in as TIFF files and run through a semi-automated analysis pipeline within ImageTank. Due to the morphologically complicated shape of A. gossypii cells, nonoverlapping hyphae were manually segmented as an initial step. For each hypha, cell volume was reconstructed using the cytoplasmic background fluorescence. A threshold was manually set to create an initial 3D mask of the hypha, followed by a morphological opening operation to remove extraneous parts of the mask caused by out-of-focus light. Nuclei were masked in 3D through a threshold followed by a size filter, where only objects larger than 1 µm3 were kept to remove DAPI-stained mitochondria.
For each RNA channel, RNA spots within a given hypha were defined as regions greater than twice the mean of that hypha’s cytoplasmic background intensity. To estimate this local background intensity, the median fluorescence within each hyphal mask was calculated. Any pixel greater than twice the median was temporarily removed from the hyphal mask. The mean fluorescence intensity within this resultant mask was used to approximate the cytoplasmic background fluorescence for each RNA channel. RNA spots were masked in 3D by applying this threshold to RNA-channel pixels within the original hyphal mask. Whether or not the center of mass of each RNA spot was within a nucleus was recorded.
Data were then exported from ImageTank and post-processed using Python within a Jupyter Notebook. Volume of the cytosol of each hypha was calculated by subtracting the volume occupied by the nuclei from the volume of the hyphal mask. Only cytoplasmic RNAs were used for calculation of concentrations. While the majority of observed RNA spots within a hypha were similar in size and intensity, some were significantly brighter than the rest, likely resulting from a higher level of transcription at that location. Furthermore, if multiple RNA spots were too close together, there is the possibility that they ended up in the same mask. To get the number of individual RNAs within a given hypha given these considerations, the median of the distribution of integrated densities of the RNA spots was used as an approximation for a single RNA. RNA spots with integrated density less than twice this median were considered to be a single RNA, and any larger than twice the median were divided by the median to estimate number of RNAs in that location. Concentration within each hypha was determined by then dividing the number of RNAs by the volume of the cytosol. Mutant strains were compared with control using ANOVA with multiple comparison correction.
Software and data availability
ImageTank can be downloaded at https://www.visualdatatools.com/ImageTank, and the smFISH image dataset and corresponding ImageTank file are available upon request.
Recombinant protein purification
Full-length Whi3 or Whi3 mutant alleles were tagged with a 6-His tag at the N terminus and transformed into BL21 bacteria for protein expression. Cultures were induced with 1 mM isopropyl β-d-1-thiogalactopyranoside after reaching OD 0.6 and grown for 20 h at 18°C. Cells were collected by centrifugation and lysed in lysis buffer (1.5 M KCl, 20 mM Tris, pH 8.0, 20 mM imidazole, pH 8, 1 mM DTT, and one tablet of Roche protease inhibitor cocktail). Lysate was centrifuged to clarify and supernatant was incubated with Ni-NTA resin. Beads were washed with lysis buffer in a gravity column and protein was eluted in elution buffer (150 mM KCl, 20 mM Tris, pH 8.0, 200 mM imidazole, pH 8.0, and 1 mM DTT). Proteins were dialyzed into droplet buffer (150 mM KCl, 20 mM Tris, pH 8.0, and 1 mM DTT) for use in phase separation assays.
In vitro RNA transcription
RNA transcription was performed as described previously (Langdon et al., 2018). Briefly, plasmid DNA was digested to obtain a linear DNA template that was used with a T7 Hiscribe in vitro transcription kit and cy3-UTP according to the manufacturer’s instructions to create Cy3-labeled versions of CLN3 and BNI1.
In vitro phase separation assays
Protein and RNA were diluted into droplet buffer in glass chambers to a final concentration of 8 µM recombinant protein and 5 nM in vitro–transcribed mRNA. Reactions were incubated at room temperature for 2 h and then imaged using a spinning disc confocal microscope (Nikon CSU-W1) with VC Plan Apo 60×/1.40 NA oil immersion objective and an sCMOS 85% QE camera (Photometrics). Quantification was performed in Fiji by using Yen AutoThreshold to create masks followed by a watershed and then the analyze particle tool in Fiji.
Online supplemental material
A representative smFISH max projection, 3D reconstruction from ImageTank, distributions of overall concentrations of cytoplasmic RNAs, and in vitro droplet assays are shown in Fig. S1. Localization and abundance of Whi3 puncta following heat stress in S289 mutants is shown in Fig. S2.
Acknowledgments
We would like to thank the Gladfelter and Lew Labs for useful discussions, and Tim Straub for consulting about statistics.
The work was supported by National Institutes of Health grant R01-GM-081506.
The authors declare no competing financial interests.
Author contributions: T.M. Gerbich conducted experiments and analyzed data. T.M. Gerbich and A.S. Gladfelter designed experiments and wrote the manuscript. G.A. McLaughlin and D. Adalsteinsson processed smFISH data. K. Cassidy and S. Gerber performed mass spectrometry.
References
- Banani S.F., Rice A.M., Peeples W.B., Lin Y., Jain S., Parker R., and Rosen M.K.. 2016. Compositional Control of Phase-Separated Cellular Bodies. Cell. 166:651–663. 10.1016/j.cell.2016.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani S.F., Lee H.O., Hyman A.A., and Rosen M.K.. 2017. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18:285–298. 10.1038/nrm.2017.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne C.P., Mitchison T.J., and Hyman A.A.. 2011. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. USA. 108:4334–4339. 10.1073/pnas.1017150108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter A.S., Hungerbuehler A.K., and Philippsen P.. 2006. Asynchronous nuclear division cycles in multinucleated cells. J. Cell Biol. 172:347–362. 10.1083/jcb.200507003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofweber M., and Dormann D.. 2019. Friend or foe-Post-translational modifications as regulators of phase separation and RNP granule dynamics. J. Biol. Chem. 294:7137–7150. 10.1074/jbc.TM118.001189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenbach A.N., Schlosser K.A., Lyons S.P., Nasa I., Gui J., Adamo M.E., and Gerber S.A.. 2018. Global assessment of its network dynamics reveals that the kinase Plk1 inhibits the phosphatase PP6 to promote Aurora A activity. Sci. Signal. 11 eaaq1441 10.1126/scisignal.aaq1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon E.M., Qiu Y., Ghanbari Niaki A., McLaughlin G.A., Weidmann C.A., Gerbich T.M., Smith J.A., Crutchley J.M., Termini C.M., Weeks K.M., et al. . 2018. mRNA structure determines specificity of a polyQ-driven phase separation. Science. 360:922–927. 10.1126/science.aar7432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Zhang H., Baker A.E., Occhipinti P., Borsuk M.E., and Gladfelter A.S.. 2013. Protein aggregation behavior regulates cyclin transcript localization and cell-cycle control. Dev. Cell. 25:572–584. 10.1016/j.devcel.2013.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Occhipinti P., and Gladfelter A.S.. 2015. PolyQ-dependent RNA-protein assemblies control symmetry breaking. J. Cell Biol. 208:533–544. 10.1083/jcb.201407105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizunuma M., Tsubakiyama R., Ogawa T., Shitamukai A., Kobayashi Y., Inai T., Kume K., and Hirata D.. 2013. Ras/cAMP-dependent protein kinase (PKA) regulates multiple aspects of cellular events by phosphorylating the Whi3 cell cycle regulator in budding yeast. J. Biol. Chem. 288:10558–10566. 10.1074/jbc.M112.402214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok J., Kim P.M., Lam H.Y., Piccirillo S., Zhou X., Jeschke G.R., Sheridan D.L., Parker S.A., Desai V., Jwa M., et al. . 2010. Deciphering protein kinase specificity through large-scale analysis of yeast phosphorylation site motifs. Sci. Signal. 3:ra12 10.1126/scisignal.2000482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair D.R., D’Ausilio C.A., Occhipinti P., Borsuk M.E., and Gladfelter A.S.. 2010. A conserved G1 regulatory circuit promotes asynchronous behavior of nuclei sharing a common cytoplasm. Cell Cycle. 9:3771–3779. 10.4161/cc.9.18.12999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz H.P., Kaufmann A., Köhli M., Laissue, P.P., and Philippsen P.. 2006. From Function to Shape: A Novel Role of a Formin in Morphogenesis of the Fungus Ashbya Gossypii In Mol. Biol. Cell. Vol. 17 pp. 130–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland J., and Walther A.. 2005. Ashbya gossypii: a model for fungal developmental biology. Nat. Rev. Microbiol. 3:421–429. 10.1038/nrmicro1148 [DOI] [PubMed] [Google Scholar]
- Zhang H., Elbaum-Garfinkle S., Langdon E.M., Taylor N., Occhipinti P., Bridges A.A., Brangwynne C.P., and Gladfelter A.S.. 2015. RNA Controls PolyQ Protein Phase Transitions. Mol. Cell. 60:220–230. 10.1016/j.molcel.2015.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
ImageTank can be downloaded at https://www.visualdatatools.com/ImageTank, and the smFISH image dataset and corresponding ImageTank file are available upon request.