Figure 5.
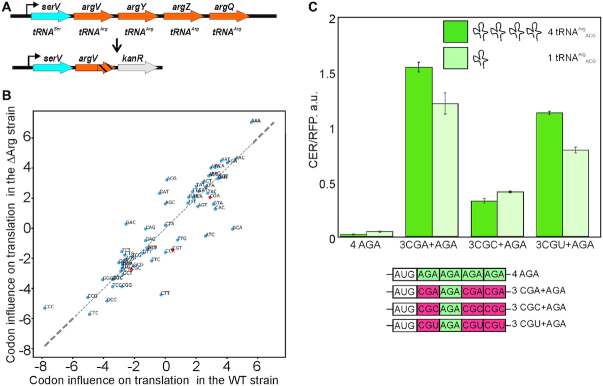
B. Influence of tRNAArgACG gene copy number on the translation of CER reporters. (A) The scheme of serV operon in the wild type and in the ΔArg strains. (B) Scatter plot of the linear regression coefficients × 103 of the triplet frequency dependences on the TEF number in the WT (x-axis) and ΔArg (y-axis) strains. Points corresponding to the arginine codons decoded by tRNAArgACG are shown in red. (C) Translation efficiency of the designed constructs. Schematic representation (lower part of the panel) and translation efficiencies (upper part of the panel) of the constructs. Translation efficiencies of the CER reporter were normalized to the reference RFP construct and indicated as a diagram. All constructs designations are indicated next to the schematic representations and below the corresponding bars. Arginine codons decoded by tRNAArgACG are shown in red, while those decoded by other tRNAs are shown in green. The sequences are listed in Supplementary Table S2. Dark green bars correspond to the wild type strain with full set of tRNAArgACG genes, while light green bars correspond to the ΔArg strain (key is provided on the graph).